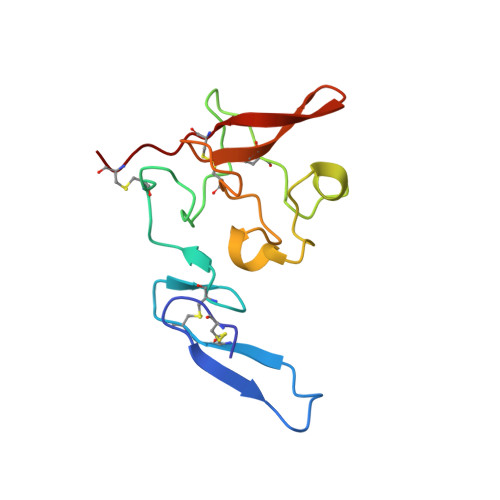
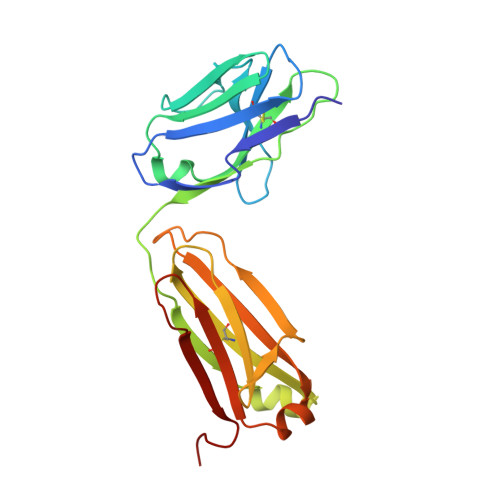
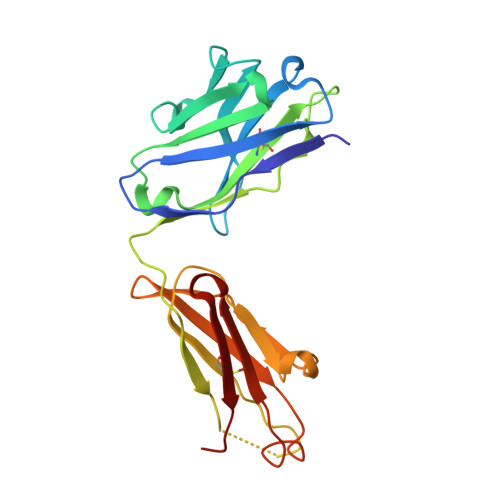
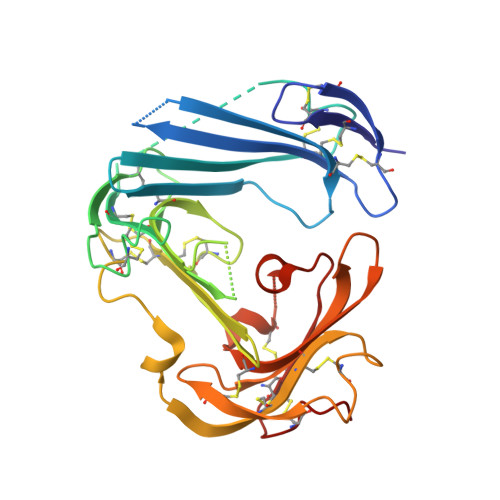
Structure of human urokinase plasminogen activator in complex with its receptor
Huai, Q., Mazar, A.P., Kuo, A., Parry, G.C., Shaw, D.E., Callahan, J., Li, Y., Yuan, C., Bian, C., Chen, L., Furie, B., Furie, B.C., Cines, D.B., Huang, M.(2006) Science 311: 656-659
- PubMed: 16456079
- DOI: https://doi.org/10.1126/science.1121143
- Primary Citation of Related Structures:
2FD6 - PubMed Abstract:
The urokinase plasminogen activator binds to its cellular receptor with high affinity and initiates signaling cascades that are implicated in pathological processes including tumor growth, metastasis, and inflammation. We report the crystal structure at 1.9 angstroms of the urokinase receptor complexed with the urokinase amino-terminal fragment and an antibody against the receptor. The three domains of urokinase receptor form a concave shape with a central cone-shaped cavity where the urokinase fragment inserts. The structure provides insight into the flexibility of the urokinase receptor that enables its interaction with a wide variety of ligands and a basis for the design of urokinase-urokinase receptor antagonists.
- Division of Hemostasis and Thrombosis, Center for Vascular Biology Research, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA 02215, USA.
Organizational Affiliation: