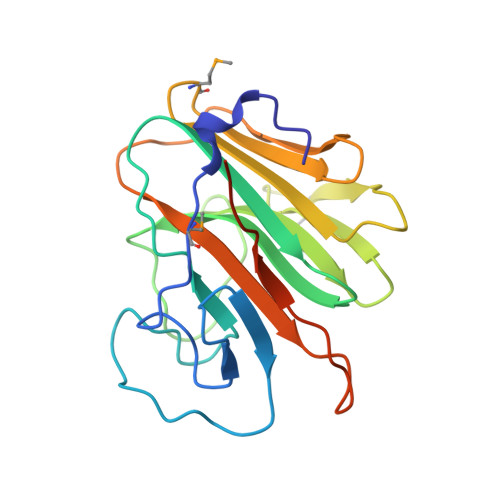
Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases
Gruetter, C., Briand, C., Capitani, G., Mittl, P.R., Papin, S., Tschopp, J., Gruetter, M.G.(2006) FEBS Lett 580: 99-106
- PubMed: 16364311
- DOI: https://doi.org/10.1016/j.febslet.2005.11.076
- Primary Citation of Related Structures:
2FBE - PubMed Abstract:
We determined the first structure of PRYSPRY, a domain found in over 500 different proteins, involved in innate immune signaling, cytokine signaling suppression, development, cell growth and retroviral restriction. The fold encompasses a 7-stranded and a 6-stranded antiparallel beta-sheet, arranged in a beta-sandwich. In the crystal, PRYSPRY forms a dimer where the C-terminus of an acceptor molecule binds to the concave surface of a donor molecule, which represents a putative interaction site. Mutations in the PRYSPRY domains of Pyrin, which are responsible for familial Mediterranean fever, map on the putative PRYSPRY interaction site.
- Department of Biochemistry, University of Zürich, CH-8057 Zürich, Switzerland.
Organizational Affiliation: