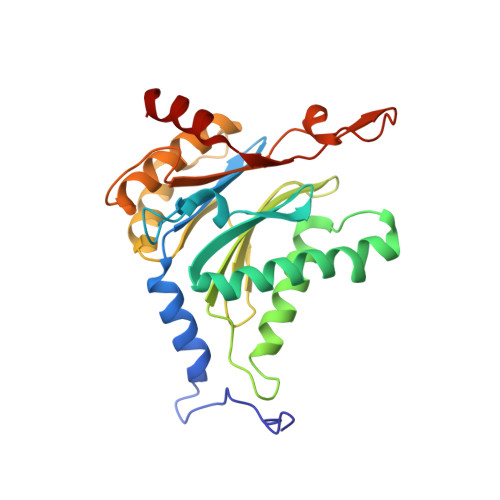
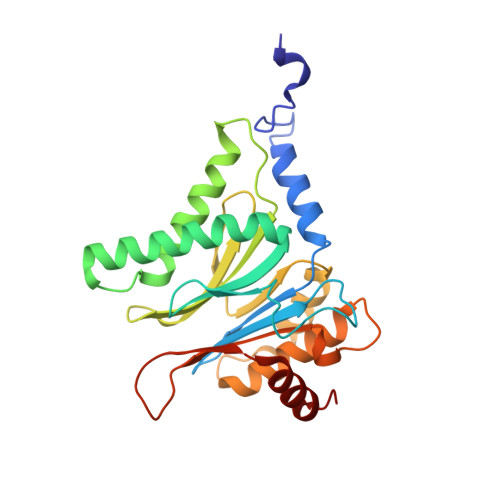
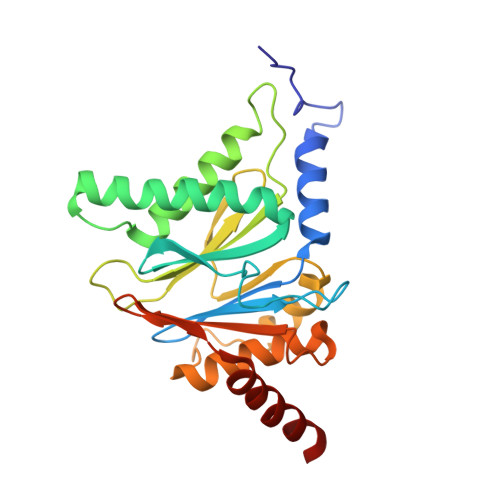
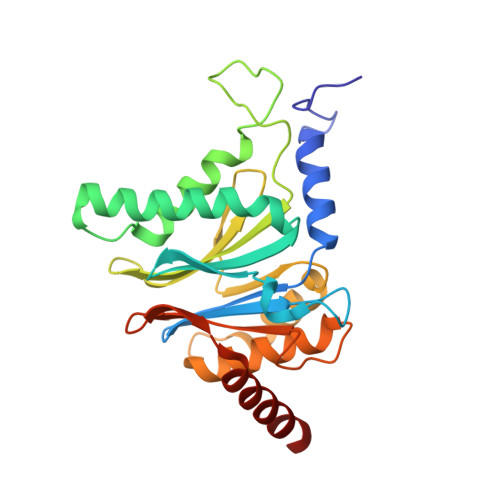
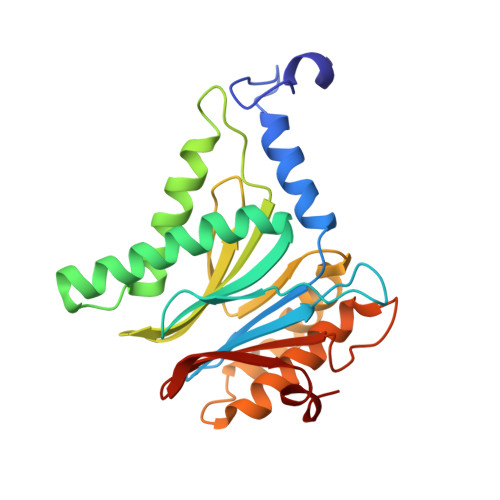
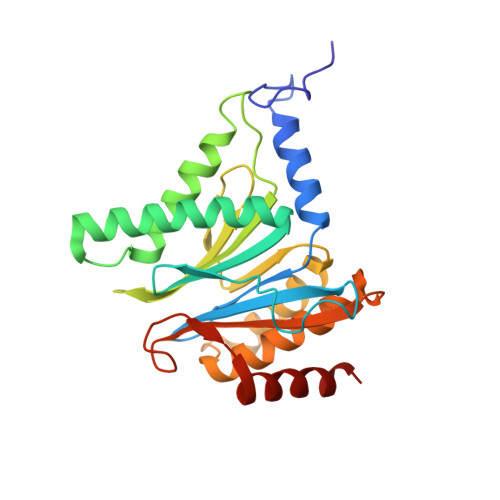
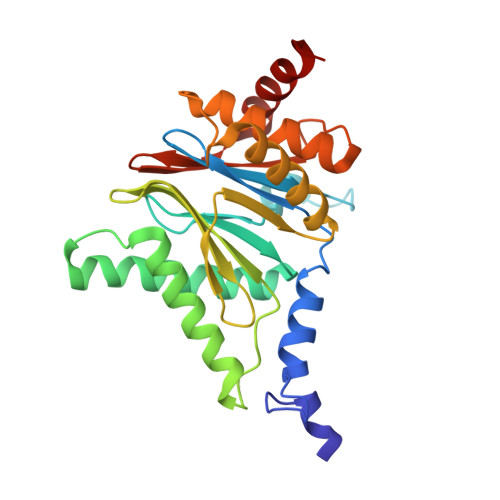
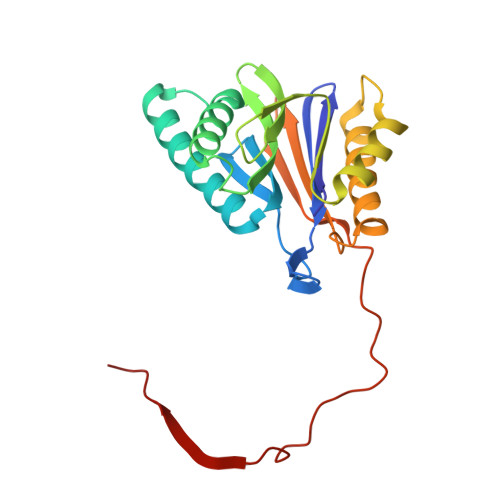
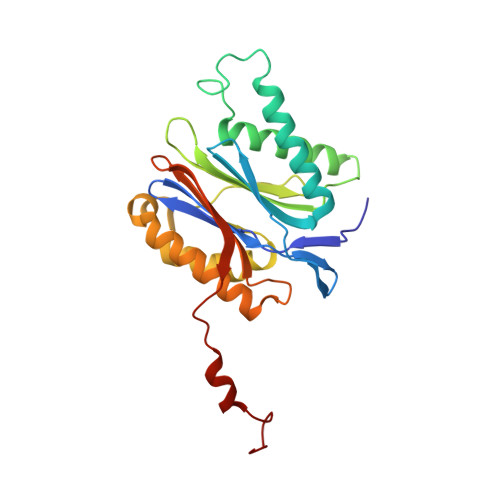
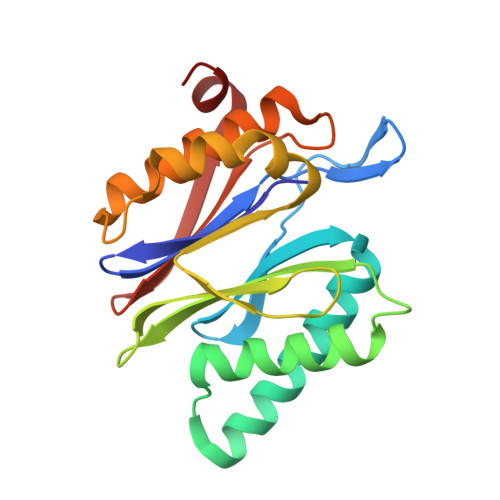
Crystal Structures of Salinosporamide A (NPI-0052) and B (NPI-0047) in Complex with the 20S Proteasome Reveal Important Consequences of beta-Lactone Ring Opening and a Mechanism for Irreversible Binding.
Groll, M., Huber, R., Potts, B.C.(2006) J Am Chem Soc 128: 5136-5141
- PubMed: 16608349
- DOI: https://doi.org/10.1021/ja058320b
- Primary Citation of Related Structures:
2FAK - PubMed Abstract:
The crystal structures of the yeast 20S proteasome core particle (CP) in complex with Salinosporamides A (NPI-0052; 1) and B (4) were solved at <3 angstroms resolution. Each ligand is covalently bound to Thr1O(gamma) via an ester linkage to the carbonyl derived from the beta-lactone ring of the inhibitor. In the case of 1, nucleophilic addition to the beta-lactone ring is followed by addition of C-3O to the chloroethyl group, giving rise to a cyclic ether. The crystal structures were compared to that of the omuralide/CP structure solved previously, and the collective data provide new insights into the mechanism of inhibition and irreversible binding of 1. Upon opening of the beta-lactone ring, C-3O assumes the position occupied by a water molecule in the unligated enzyme and hinders deacylation of the enzyme-ligand complex. Furthermore, the resulting protonation state of Thr1NH2 deactivates the catalytic N-terminus.
- Ludwig-Maximilians-University of Munich, Butenandtstr. 5, Building B, 81377 Munich, Germany. Michael.Groll@med.uni-muenchen.de
Organizational Affiliation: