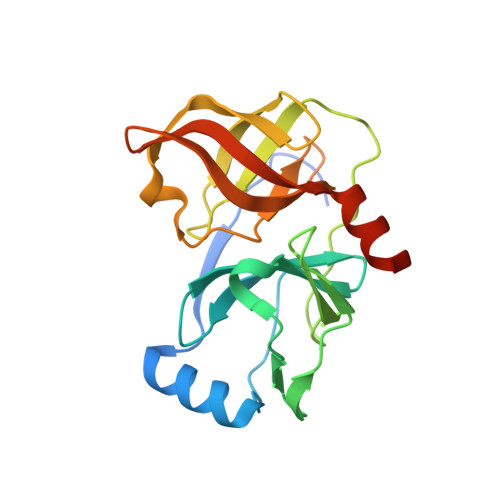
Depeptidization efforts on P3-P2 a-ketoamide inhibitors of HCV NS3-4A serine protease: Effect on HCV replicon activity.
Bogen, S.L., Ruan, S., Liu, R., Agrawal, S., Pichardo, J., Prongay, A., Baroudy, B., Saksena, A., Girijavallabhan, V., Njoroge, F.G.(2006) Bioorg Med Chem Lett 16: 1621-1627
- PubMed: 16387495
- DOI: https://doi.org/10.1016/j.bmcl.2005.12.013
- Primary Citation of Related Structures:
2F9V - PubMed Abstract:
Depeptidization efforts of the P(3)-P(2) region of P(3) capped alpha-ketoamide inhibitor of HCV NS3 serine protease 1 are reported. We clearly established that N-methylation of the P(2) nitrogen and modification of the P(2)' carboxylic acid terminus were essential for activity in the replicon assay.
- Schering-Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, NJ 07033, USA. stephane.bogen@spcorp.com
Organizational Affiliation: