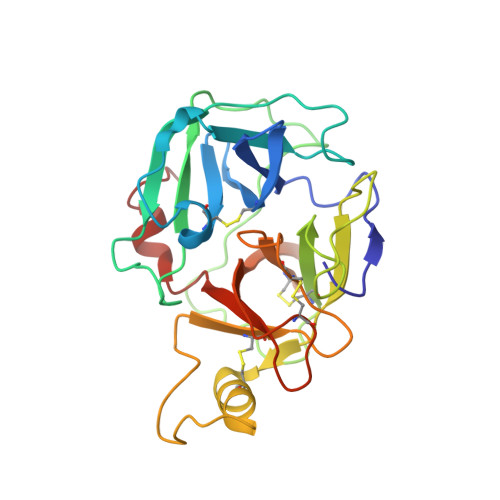
X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
Rohr, K.B., Selwood, T., Marquardt, U., Huber, R., Schechter, N.M., Bode, W., Than, M.E.(2005) J Mol Biology 357: 195-209
- PubMed: 16414069
- DOI: https://doi.org/10.1016/j.jmb.2005.12.037
- Primary Citation of Related Structures:
2F9N, 2F9O, 2F9P - PubMed Abstract:
Tryptases alpha and beta are trypsin-like serine proteinases expressed in large amounts by mast cells. Beta-tryptase is a tetramer that has enzymatic activity, but requires heparin binding to maintain functional and structural stability, whereas alpha-tryptase has little, if any, enzymatic activity but is a stable tetramer in the absence of heparin. As shown previously, these differences can be mainly attributed to the different conformations of the 214-220 segment. Interestingly, the replacement of Asp216 by Gly, which is present in beta-tryptase, results in enzymatically active but less stable alpha-tryptase mutants. We have solved the crystal structures of both the single (D216G) and the double (K192Q/D216G) mutant forms of recombinant human alphaI-tryptase in complex with the peptide inhibitor leupeptin, as well as the structure of the non-inhibited single mutant. The inhibited mutants exhibited an open functional substrate binding site, while in the absence of an inhibitor, the open (beta-tryptase-like) and the closed (alpha-tryptase-like) conformations were present simultaneously. This shows that both forms are in a two-state equilibrium, which is influenced by the residues in the vicinity of the active site and by inhibitor/substrate binding. Novel insights regarding the observed stability differences as well as a potential proteolytic activity of wild-type alpha-tryptase, which may possess a cryptic active site, are discussed.
- Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: