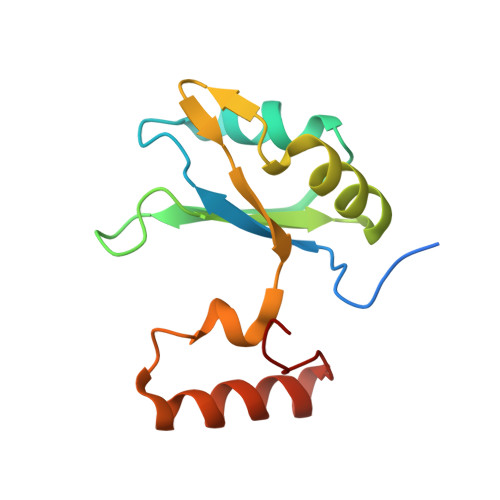
Crystal structure of a core spliceosomal protein interface
Schellenberg, M.J., Edwards, R.A., Ritchie, D.B., Kent, O.A., Golas, M.M., Stark, H., Glover, J.N.M., Macmillan, A.M.(2006) Proc Natl Acad Sci U S A 103: 1266-1271
- PubMed: 16432215
- DOI: https://doi.org/10.1073/pnas.0508048103
- Primary Citation of Related Structures:
2F9D, 2F9J - PubMed Abstract:
The precise excision of introns from precursor mRNAs (pre-mRNAs) in eukaryotes is accomplished by the spliceosome, a complex assembly containing five small nuclear ribonucleoprotein (snRNP) particles. Human p14, a component of the spliceosomal U2 and U11/U12 snRNPs, has been shown to associate directly with the pre-mRNA branch adenosine early in spliceosome assembly and within the fully assembled spliceosome. Here we report the 2.5-A crystal structure of a complex containing p14 and a peptide derived from the p14-associated U2 snRNP component SF3b155. p14 contains an RNA recognition motif (RRM), the surface of which is largely occluded by a C-terminal alpha-helix and a portion of the SF3b155 peptide. An analysis of RNA.protein crosslinking to wild-type and mutant p14 shows that the branch adenosine directly interacts with a conserved aromatic within a pocket on the surface of the complex. This result, combined with a comparison of the structure with known RRMs and pseudoRRMs as well as model-building by using the electron cryomicroscopy structure of a spliceosomal U11/U12 di-snRNP, suggests that p14.SF3b155 presents a noncanonical surface for RNA recognition at the heart of the mammalian spliceosome.
- Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada T6G 2H7.
Organizational Affiliation: