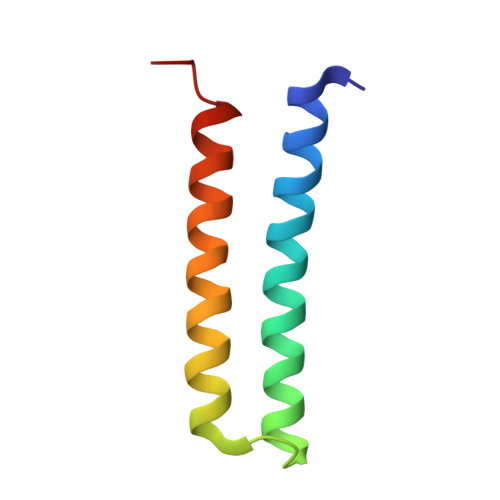
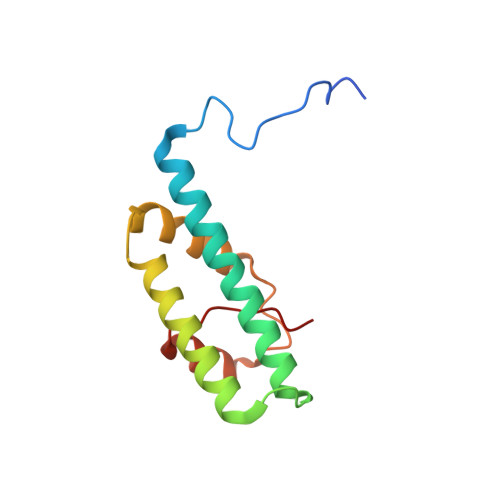
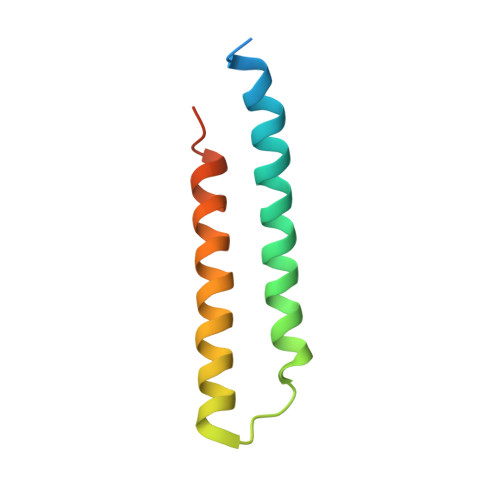
Structural and functional organization of the ESCRT-I trafficking complex.
Kostelansky, M.S., Sun, J., Lee, S., Kim, J., Ghirlando, R., Hierro, A., Emr, S.D., Hurley, J.H.(2006) Cell 125: 113-126
- PubMed: 16615894
- DOI: https://doi.org/10.1016/j.cell.2006.01.049
- Primary Citation of Related Structures:
2F66, 2F6M - PubMed Abstract:
The endosomal sorting complex required for transport (ESCRT) complexes are central to receptor downregulation, lysosome biogenesis, and budding of HIV. The yeast ESCRT-I complex contains the Vps23, Vps28, and Vps37 proteins, and its assembly is directed by the C-terminal steadiness box of Vps23, the N-terminal half of Vps28, and the C-terminal half of Vps37. The crystal structures of a Vps23:Vps28 core subcomplex and the Vps23:Vps28:Vps37 core were solved at 2.1 and 2.8 A resolution. Each subunit contains a structurally similar pair of helices that form the core. The N-terminal domain of Vps28 has a hydrophobic binding site on its surface that is conformationally dynamic. The C-terminal domain of Vps28 binds the ESCRT-II complex. The structure shows how ESCRT-I is assembled by a compact core from which the Vps23 UEV domain, the Vps28 C domain, and other domains project to bind their partners.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, US Department of Health and Human Services, Bethesda, MD 20892, USA.
Organizational Affiliation: