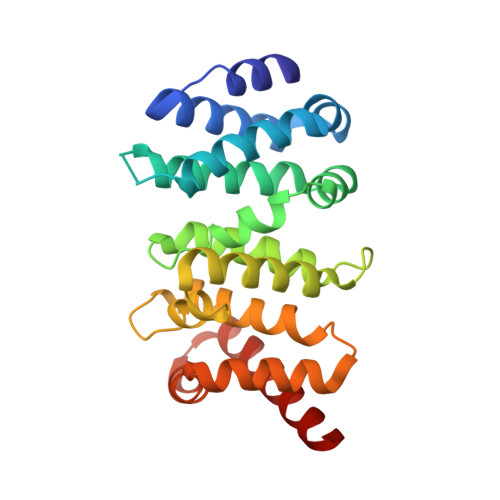
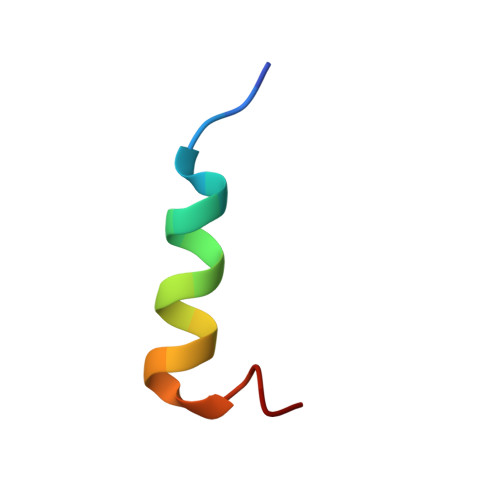
Structure of the Autoinhibitory Switch in Formin mDia1
Nezami, A.G., Poy, F., Eck, M.J.(2006) Structure 14: 257-263
- PubMed: 16472745
- DOI: https://doi.org/10.1016/j.str.2005.12.003
- Primary Citation of Related Structures:
2F31 - PubMed Abstract:
Diaphanous-related formins (DRFs) regulate the nucleation and polymerization of unbranched actin filaments. The activity of DRFs is inhibited by an intramolecular interaction between their N-terminal regulatory region and a conserved C-terminal segment termed the Diaphanous autoinhibitory domain (DAD). Binding of GTP bound Rho to the mDia1 N terminus releases this autoinhibitory restraint. Here, we describe the crystal structure of the DAD segment of mDia1 in complex with the relevant N-terminal fragment, termed the DID domain. The structure reveals that the DAD segment forms an amphipathic helix that binds a conserved, concave surface on the DID domain. Comparison with the structure of the mDia1 N terminus bound to RhoC suggests that release of the autoinhibitory DAD interaction is accomplished largely by Rho-induced restructuring of the adjacent GTPase binding subdomain (GBD), but also by electrostatic repulsion and a small, direct steric occlusion of the DAD binding cleft by Rho itself.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 44 Binney Street, Boston, Massachusetts 02115, USA.
Organizational Affiliation: