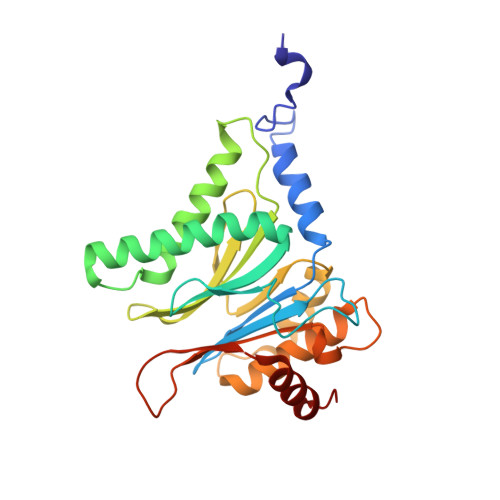
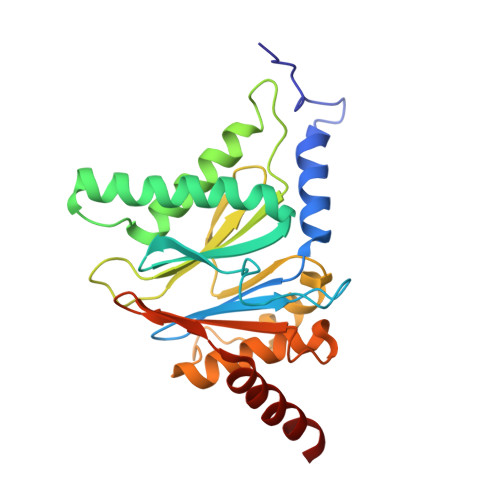
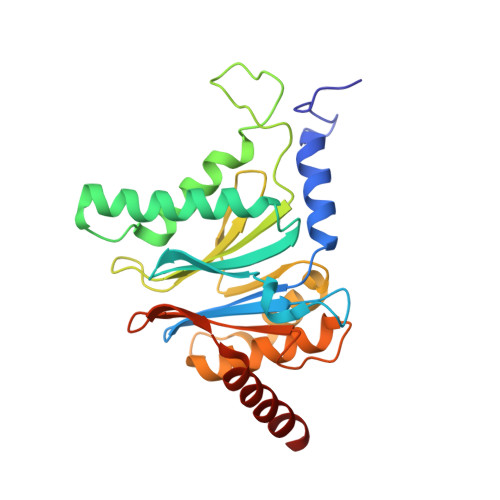
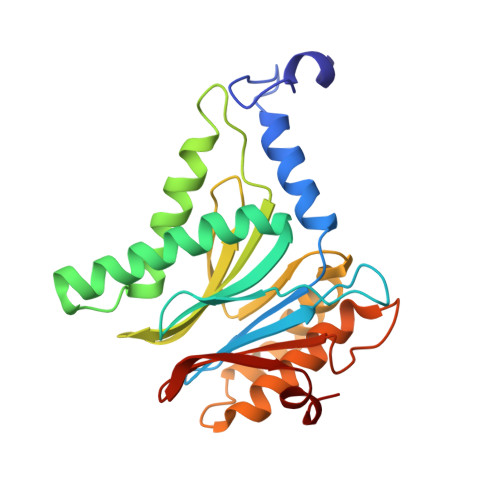
Crystal Structure of the Boronic Acid-Based Proteasome Inhibitor Bortezomib in Complex with the Yeast 20S Proteasome.
Groll, M., Berkers, C.R., Ploegh, H.L., Ovaa, H.(2006) Structure 14: 451-456
- PubMed: 16531229
- DOI: https://doi.org/10.1016/j.str.2005.11.019
- Primary Citation of Related Structures:
2F16 - PubMed Abstract:
The dipeptide boronic acid bortezomib, also termed VELCADE, is a proteasome inhibitor now in use for the treatment of multiple myeloma, and its use for the treatment of other malignancies is being explored. We determined the crystal structure of the yeast 20S proteasome in complex with bortezomib to establish the specificity and binding mode of bortezomib to the proteasome's different catalytically active sites. This structure should enable the rational design of new boronic acid derivatives with improved affinities and specificities for individual active subunits.
- Department for Physiological Chemistry, Ludwig-Maximilians-University, Butenandtstrasse 5, Building B 81377, München, Germany. mgroll@med.uni-muenchen.de
Organizational Affiliation: