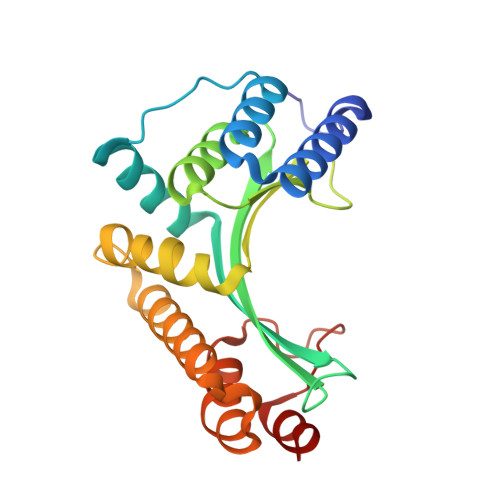
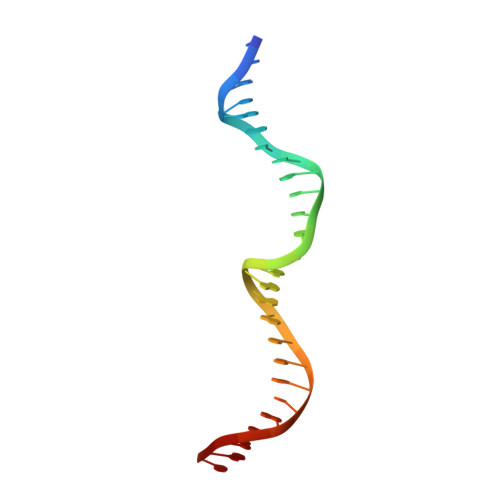
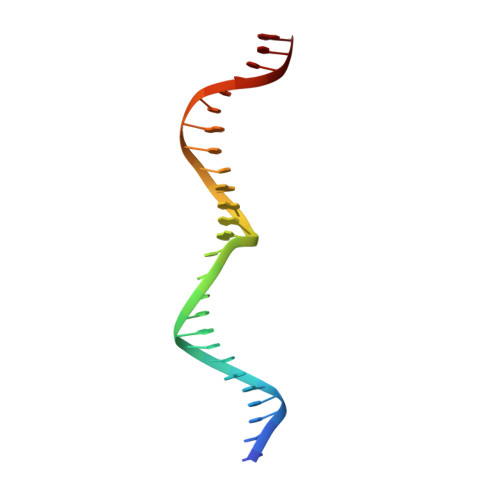
The structure of I-CeuI homing endonuclease: Evolving asymmetric DNA recognition from a symmetric protein scaffold.
Spiegel, P.C., Chevalier, B., Sussman, D., Turmel, M., Lemieux, C., Stoddard, B.L.(2006) Structure 14: 869-880
- PubMed: 16698548
- DOI: https://doi.org/10.1016/j.str.2006.03.009
- Primary Citation of Related Structures:
2EX5 - PubMed Abstract:
Homing endonucleases are highly specific catalysts of DNA strand breaks, leading to the transfer of mobile intervening sequences containing the endonuclease ORF. We have determined the structure and DNA recognition behavior of I-CeuI, a homodimeric LAGLIDADG endonuclease from Chlamydomonas eugametos. This symmetric endonuclease displays unique structural elaborations on its core enzyme fold, and it preferentially cleaves a highly asymmetric target site. This latter property represents an early step, prior to gene fusion, in the generation of asymmetric DNA binding platforms from homodimeric ancestors. The divergence of the sequence, structure, and target recognition behavior of homing endonucleases, as illustrated by this study, leads to the invasion of novel genomic sites by mobile introns during evolution.
- Graduate Programs in Biomolecular Structure and Design and Molecular and Cellular Biology, University of Washington, Seattle, Washington 98195, USA.
Organizational Affiliation: