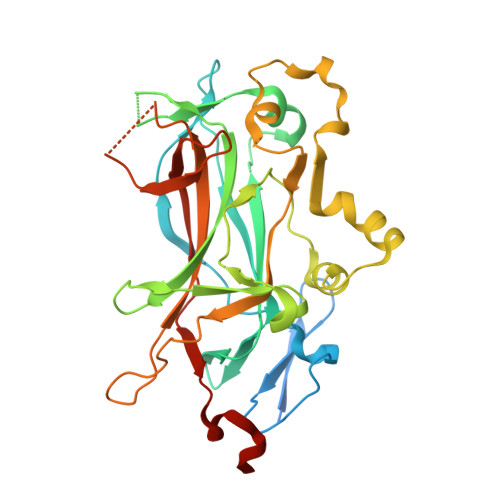
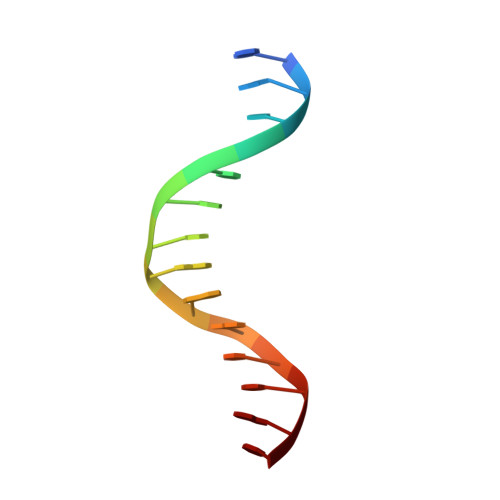
Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Lamoureux, J.S., Glover, J.N.(2006) Structure 14: 555-565
- PubMed: 16531239
- DOI: https://doi.org/10.1016/j.str.2005.11.017
- Primary Citation of Related Structures:
2ETW, 2EUV, 2EUW, 2EUX, 2EUZ, 2EVF, 2EVG, 2EVH, 2EVI, 2EVJ - PubMed Abstract:
The Saccharomyces cerevisiae transcription factor Ndt80 selectively binds a DNA consensus sequence (the middle sporulation element [MSE]) to activate gene expression after the successful completion of meiotic recombination. Here we report the X-ray crystal structures of Ndt80 bound to ten distinct MSE variants. Comparison of these structures with the structure of Ndt80 bound to a consensus MSE reveals structural principles that determine the DNA binding specificity of this transcription factor. The 5' GC-rich end of the MSE contains distinct 5'-YpG-3' steps that are recognized by arginine side chains through a combination of hydrogen bonding and cation-pi interactions. The 3' AT-rich region is recognized via minor groove contacts that sterically exclude the N2 atom of GC base pairs. The conformation of the AT-rich region is fixed by interactions with the protein that favor recognition of poly(A)-poly(T) versus mixed AT sequences through an avoidance of major groove steric clashes at 5'-ApT-3' steps.
- Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.
Organizational Affiliation: