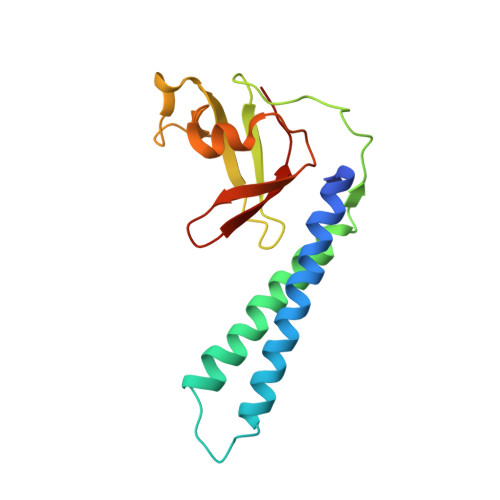
Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage.
Lamour, V., Hogan, B.P., Erie, D.A., Darst, S.A.(2006) J Mol Biology 356: 179-188
- PubMed: 16337964
- DOI: https://doi.org/10.1016/j.jmb.2005.10.083
- Primary Citation of Related Structures:
2ETN - PubMed Abstract:
Transcription elongation in bacteria is promoted by Gre-factors, which stimulate an endogenous, endonucleolytic transcript cleavage activity of the RNA polymerase. A GreA paralog, Gfh1, present in Thermus aquaticus and Thermus thermophilus, has the opposite effect on elongation complexes, inhibiting rather than stimulating transcript cleavage. We have determined the 3.3 angstroms-resolution X-ray crystal structure of T.aquaticus Gfh1. The structure reveals an N-terminal and a C-terminal domain with close structural similarity to the domains of GreA, but with an unexpected conformational change in terms of the orientation of the domains with respect to each other. However, structural and functional analysis suggests that when complexed with RNA polymerase, Gfh1 adopts a conformation similar to that of GreA. These results reveal considerable structural flexibility for Gfh1, and for Gre-factors in general, as suggested by structural modeling, and point to a possible role for the conformational switch in Gre-factor and Gfh1 regulation. The opposite functional effect of Gfh1 compared with GreA may be determined by three structural characteristics. First, Gfh1 lacks the basic patch present in Gre-factors that likely plays a role in anchoring the 3'-fragment of the back-tracked RNA. Second, the loop at the tip of the N-terminal coiled-coil is highly flexible and contains extra acidic residues compared with GreA. Third, the N-terminal coiled-coil finger lacks a kink in the first alpha-helix, resulting in a straight coiled-coil compared with GreA. The latter two characteristics suggest that Gfh1 chelates a magnesium ion in the RNA polymerase active site (like GreA) but in a catalytically inactive configuration.
- The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: