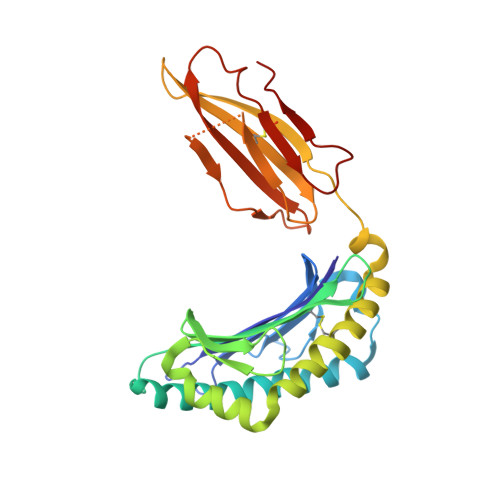
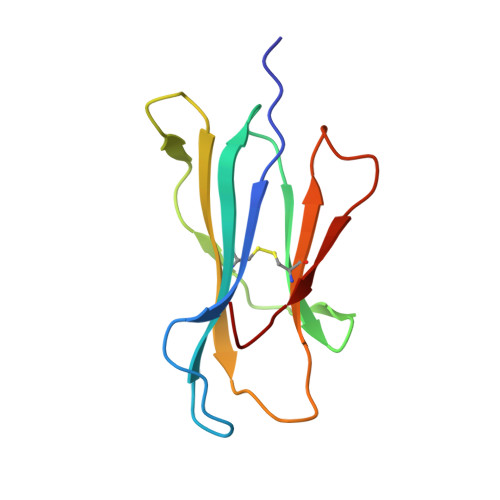
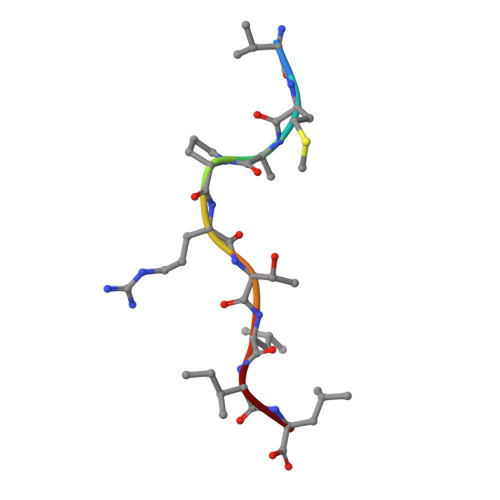
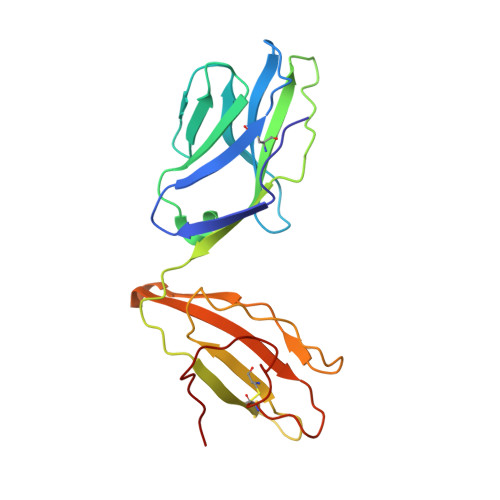
Structural basis for a major histocompatibility complex class Ib-restricted T cell response
Hoare, H.L., Sullivan, L.C., Pietra, G., Clements, C.S., Lee, E.J., Ely, L.K., Beddoe, T., Falco, M., Kjer-Nielsen, L., Reid, H.H., McCluskey, J., Moretta, L., Rossjohn, J., Brooks, A.G.(2006) Nat Immunol 7: 256-264
- PubMed: 16474394
- DOI: https://doi.org/10.1038/ni1312
- Primary Citation of Related Structures:
2ESV - PubMed Abstract:
In contrast to antigen-specific immunity orchestrated by major histocompatibility complex (MHC) class Ia molecules, the ancestrally related nonclassical MHC class Ib molecules generally mediate innate immune responses. Here we have demonstrated the structural basis by which the MHC class Ib molecule HLA-E mediates an adaptive MHC-restricted cytotoxic T lymphocyte response to human cytomegalovirus. Highly constrained by host genetics, the response showed notable fine specificity for position 8 of the viral peptide, which is the sole discriminator of self versus nonself. Despite the evolutionary divergence of MHC class Ia and class Ib molecules, the structure of the T cell receptor-MHC class Ib complex was very similar to that of conventional T cell receptor-MHC class Ia complexes. These results emphasize the evolutionary 'ambiguity' of HLA-E, which not only interacts with innate immune receptors but also has the functional capacity to mediate virus-specific cytotoxic T lymphocyte responses during adaptive immunity.
- The Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: