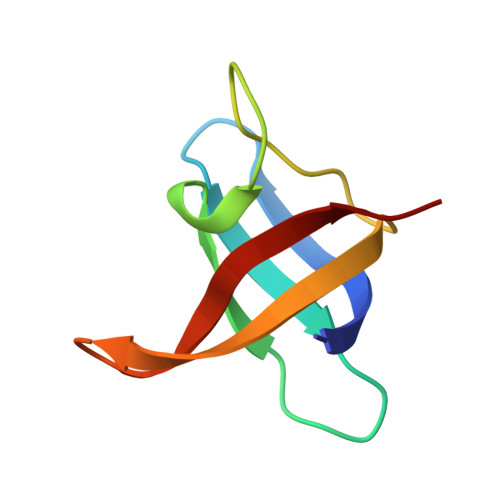
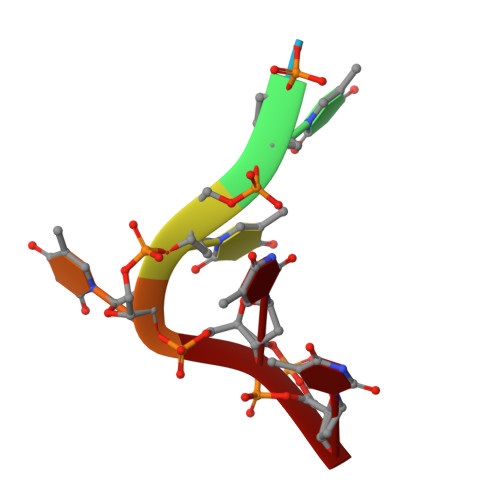
T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB.
Max, K.E., Zeeb, M., Bienert, R., Balbach, J., Heinemann, U.(2006) J Mol Biology 360: 702-714
- PubMed: 16780871
- DOI: https://doi.org/10.1016/j.jmb.2006.05.044
- Primary Citation of Related Structures:
2ES2 - PubMed Abstract:
Bacterial cold shock proteins (CSPs) are involved in cellular adaptation to cold stress. They bind to single-stranded nucleic acids with a KD value in the micro- to nanomolar range. Here we present the structure of the Bacillus subtilis CspB (Bs-CspB) in complex with hexathymidine (dT6) at a resolution of 1.78 A. Bs-CspB binds to dT6 with nanomolar affinity via an amphipathic interface on the protein surface. Individual binding subsites interact with single nucleobases through stacking interactions and hydrogen bonding. The sugar-phosphate backbone and the methyl groups of the thymine nucleobases remain solvent exposed and are not contacted by protein groups. Fluorescence titration experiments monitoring the binding of oligopyrimidines to Bs-CspB reveal binding preferences at individual subsites and allow the design of an optimised heptapyrimidine ligand, which is bound with sub-nanomolar affinity. This study reveals the stoichiometry and sequence determinants of the binding of single-stranded nucleic acids to a preformed site on Bs-CspB and thus provides the structural basis of the RNA chaperone and transcription antitermination activities of the CSP.
- Max-Delbrück-Centrum für Molekulare Medizin 13125 Berlin, Germany.
Organizational Affiliation: