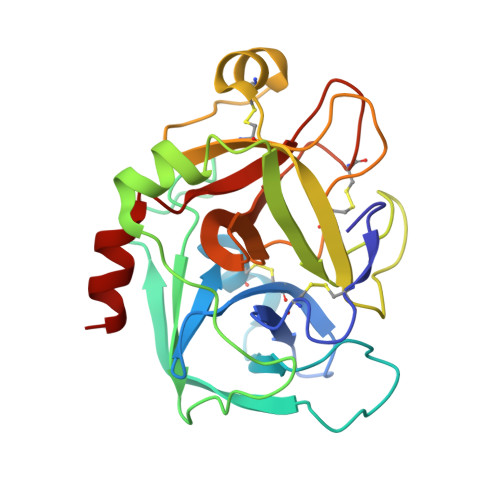
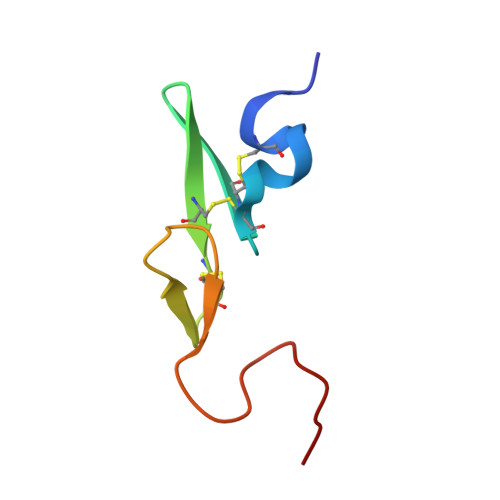
Discovery of N-{(1R,2S,5S)-2-{[(5-Chloroindol-2-yl)carbonyl]amino}-5-[(dimethylamino)carbonyl]cyclohexyl}-5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine-2-carboxamide hydrochloride: A Novel, Potent and Orally Active Direct Inhibitor of Factor Xa
Nagata, T., Yoshino, T., Haginoya, N., Yoshikawa, K., Nagamochi, M., Kobayashi, S., Komoriya, S., Yokomizo, A., Muto, R., Yamaguchi, K., Osanai, K., Suzuki, M., Kanno, H.To be published.