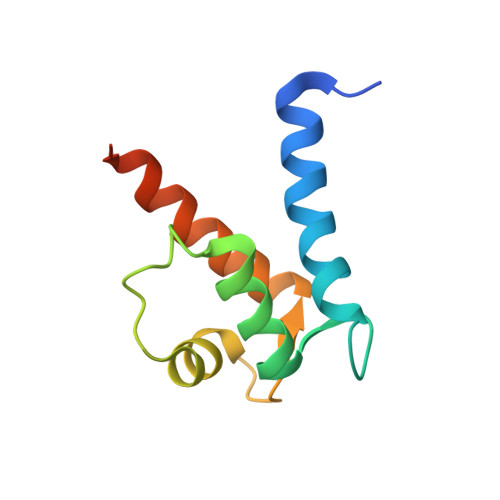
Crystal structure of human S100A13 in the Ca2+-bound state
Imai, F.L., Nagata, K., Yonezawa, N., Nakano, M., Tanokura, M.(2008) Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 70-76
- PubMed: 18259052
- DOI: https://doi.org/10.1107/S1744309107068236
- Primary Citation of Related Structures:
2EGD - PubMed Abstract:
S100A13 is a member of the S100 family of EF-hand-containing calcium-binding proteins. S100A13 plays an important role in the secretion of fibroblast growth factor-1 and interleukin 1 alpha, two pro-angiogenic factors released by the nonclassical endoplasmic reticulum/Golgi-independent secretory pathway. The X-ray crystal structure of human S100A13 at pH 7.5 was determined at 1.8 A resolution. The structure was solved by molecular replacement and was refined to a final R factor of 19.0%. The structure revealed that human S100A13 exists as a homodimer with two calcium ions bound to each protomer. The protomer is composed of four alpha-helices (alpha(1)-alpha(4)), which form a pair of EF-hand motifs. Dimerization occurs by hydrophobic interactions between helices alpha(1) and alpha(4) and by intermolecular hydrogen bonds between residues from helix alpha(1) and the residues between alpha(2) and alpha(3) of both chains. Despite the high similarity of the backbone conformation in each protomer, the crystal structures of human S100A13 at pH 7.5 (this study) and at pH 6.0 [Li et al. (2007), Biochem. Biophys. Res. Commun. 356, 616-621] exhibit recognizable differences in the relative orientation ( approximately 2.5 degrees) of the protomers within the dimer and also remarkable differences in the side-chain conformations of several amino-acid residues.
- Department of Chemistry, Graduate School of Science, Chiba University, 1-33 Yayoi-cho, Inage-ku, Chiba 263-8522, Japan.
Organizational Affiliation: