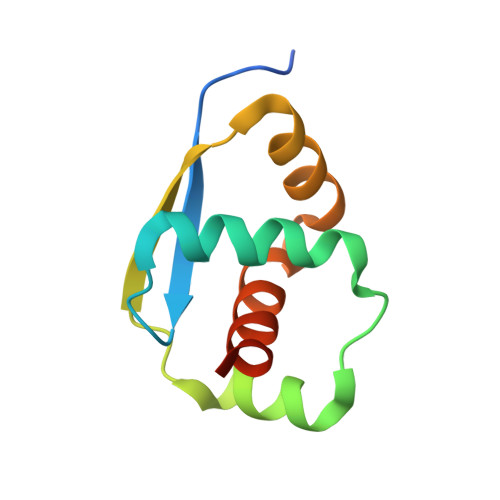
Crystal structure analysis of a hypothetical protein (MJ0366) from Methanocaldococcus jannaschii revealed a novel topological arrangement of the knot fold
Thiruselvam, V., Kumarevel, T., Karthe, P., Kuramitsu, S., Yokoyama, S., Ponnuswamy, M.N.(2017) Biochem Biophys Res Commun 482: 264-269
- PubMed: 27845039
- DOI: https://doi.org/10.1016/j.bbrc.2016.11.052
- Primary Citation of Related Structures:
2EFV - PubMed Abstract:
The crystal structure of a hypothetical protein MJ0366, derived from Methanocaldococcus jannaschii was solved at 1.9 Å resolution using synchrotron radiation. MJ0366 was crystallized as a monomer and has knot structural arrangement. Intriguingly, the solved structure consists of novel 'KNOT' fold conformation. The 3 1 trefoil knot was observed in the structure. The N-terminal and C-terminal ends did not participate in knot formation.
- Centre of Advanced Study in Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai, 600025, India.
Organizational Affiliation: