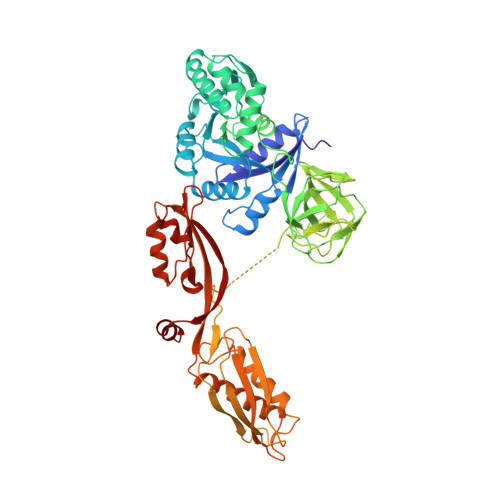
The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
Czworkowski, J., Wang, J., Steitz, T.A., Moore, P.B.(1994) EMBO J 13: 3661-3668
- PubMed: 8070396
- DOI: https://doi.org/10.1002/j.1460-2075.1994.tb06675.x
- Primary Citation of Related Structures:
1EFG, 2EFG - PubMed Abstract:
Elongation factor G (EF-G) catalyzes the translocation step of protein synthesis in bacteria, and like the other bacterial elongation factor, EF-Tu--whose structure is already known--it is a member of the GTPase superfamily. We have determined the crystal structure of EF-G--GDP from Thermus thermophilus. It is an elongated molecule whose large, N-terminal domain resembles the G domain of EF-Tu, except for a 90 residue insert, which covers a surface that is involved in nucleotide exchange in EF-Tu and other G proteins. The tertiary structures of the second domains of EF-G and EF-Tu are nearly identical, but the relative placement of the first two domains in EF-G--GDP resembles that seen in EF-Tu--GTP, not EF-Tu--GDP. The remaining three domains of EF-G look like RNA binding domains, and have no counterparts in EF-Tu.
- Department of Chemistry, Yale University, New Haven, CT 06520.
Organizational Affiliation: