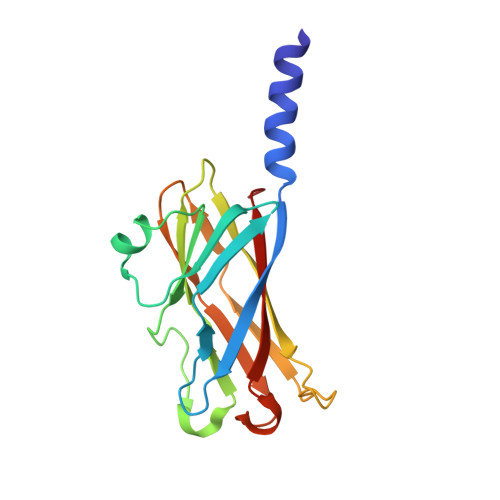
Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship
Tang, X., Wu, J., Sivaraman, J., Hew, C.L.(2007) J Virol 81: 6709-6717
- PubMed: 17409146
- DOI: https://doi.org/10.1128/JVI.02505-06
- Primary Citation of Related Structures:
2ED6, 2EDM - PubMed Abstract:
White spot syndrome virus (WSSV) is a virulent pathogen known to infect various crustaceans. It has bacilliform morphology with a tail-like appendage at one end. The envelope consists of four major proteins. Envelope structural proteins play a crucial role in viral infection and are believed to be the first molecules to interact with the host. Here, we report the localization and crystal structure of major envelope proteins VP26 and VP28 from WSSV at resolutions of 2.2 and 2.0 A, respectively. These two proteins alone account for approximately 60% of the envelope, and their structures represent the first two structural envelope proteins of WSSV. Structural comparisons among VP26, VP28, and other viral proteins reveal an evolutionary relationship between WSSV envelope proteins and structural proteins from other viruses. Both proteins adopt beta-barrel architecture with a protruding N-terminal region. We have investigated the localization of VP26 and VP28 using immunoelectron microscopy. This study suggests that VP26 and VP28 are located on the outer surface of the virus and are observed as a surface protrusion in the WSSV envelope, and this is the first convincing observation for VP26. Based on our studies combined with the literature, we speculate that the predicted N-terminal transmembrane region of VP26 and VP28 may anchor on the viral envelope membrane, making the core beta-barrel protrude outside the envelope, possibly to interact with the host receptor or to fuse with the host cell membrane for effective transfer of the viral infection. Furthermore, it is tempting to extend this host interaction mode to other structural viral proteins of similar structures. Our finding has the potential to extend further toward drug and vaccine development against WSSV.
- Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543, Republic of Singapore.
Organizational Affiliation: