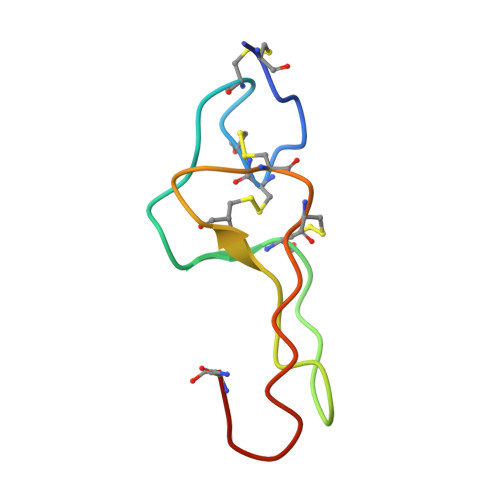
Echistatin: the refined structure of a disintegrin in solution by 1H NMR and restrained molecular dynamics.
Atkinson, R.A., Saudek, V., Pelton, J.T.(1994) Int J Pept Protein Res 43: 563-572
- PubMed: 7928087
- DOI: https://doi.org/10.1111/j.1399-3011.1994.tb00558.x
- Primary Citation of Related Structures:
2ECH - PubMed Abstract:
The structure of the disintegrin echistatin has been determined by 1H NMR, distance geometry calculations and restrained molecular dynamics simulations. The structure has been refined from the preliminary distance geometry calculations with the inclusion of additional 1H NMR data and hydrogen bonds identified in early stages of the molecular dynamics calculations. The calculations reported here allow a distinction to be made between the two possible disulfide bridging patterns-echistatin is crosslinked as follows: Cys2-Cys11, Cys7-Cys32, Cys8-Cys37, Cys20-Cys39. The final set of structures gives an average pairwise root mean square distance of 0.100 nm (calculated over the backbone atoms of residues Ser4-Cys20 and Asp30-Pro40). The core of echistatin is a well defined though irregular structure, composed of a series of non-classical turns crosslinked by the disulfide bridges and stabilised by hydrogen bonds. The RGD sequence is located in a protruding loop whose stem is formed by two rigid, hydrogen-bonded strands (Thr18-Cys20, Asp30-Cys32). The RGD sequence is connected to this structure by short, flexible segments. High (but not unlimited) mobility is probably necessary for fast recognition and fitting to the integrin receptors. Sequence variability among the disintegrins is found in the segments flanking the RGD sequence, suggesting that these may be important in conferring specificity for the receptors.
- Marion Merrell Dow Research Institute, Strasbourg, France.
Organizational Affiliation: