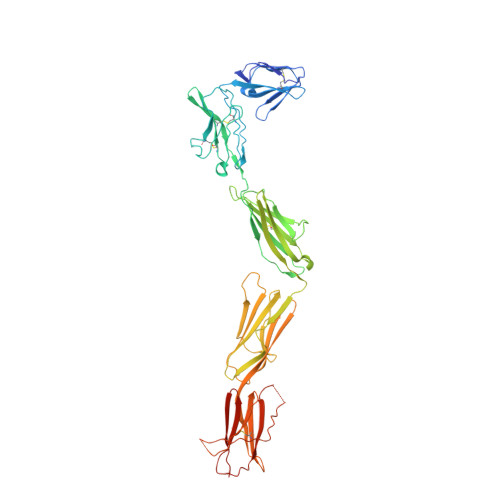
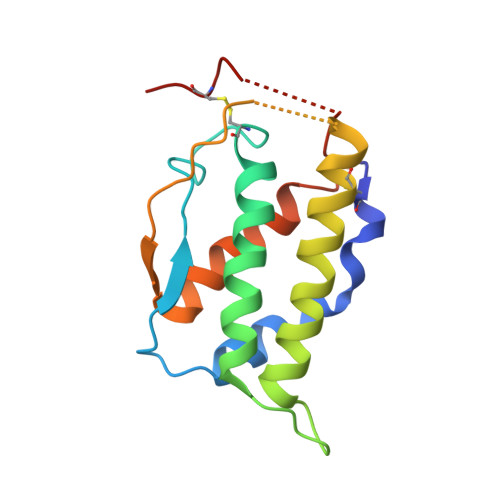
Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Yuzawa, S., Opatowsky, Y., Zhang, Z., Mandiyan, V., Lax, I., Schlessinger, J.(2007) Cell 130: 323-334
- PubMed: 17662946
- DOI: https://doi.org/10.1016/j.cell.2007.05.055
- Primary Citation of Related Structures:
2E9W, 2EC8 - PubMed Abstract:
Stem Cell Factor (SCF) initiates its multiple cellular responses by binding to the ectodomain of KIT, resulting in tyrosine kinase activation. We describe the crystal structure of the entire ectodomain of KIT before and after SCF stimulation. The structures show that KIT dimerization is driven by SCF binding whose sole role is to bring two KIT molecules together. Receptor dimerization is followed by conformational changes that enable lateral interactions between membrane proximal Ig-like domains D4 and D5 of two KIT molecules. Experiments with cultured cells show that KIT activation is compromised by point mutations in amino acids critical for D4-D4 interaction. Moreover, a variety of oncogenic mutations are mapped to the D5-D5 interface. Since key hallmarks of KIT structures, ligand-induced receptor dimerization, and the critical residues in the D4-D4 interface, are conserved in other receptors, the mechanism of KIT stimulation unveiled in this report may apply for other receptor activation.
- Department of Pharmacology, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520, USA.
Organizational Affiliation: