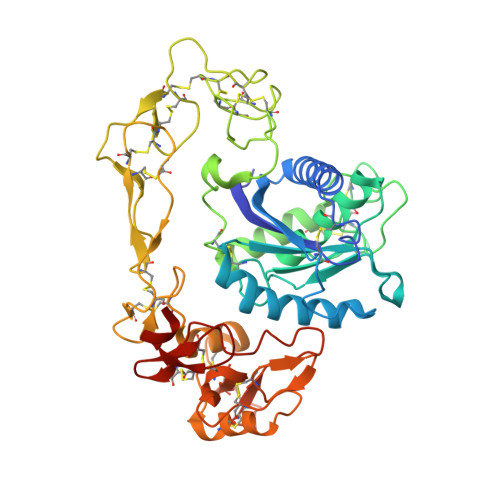
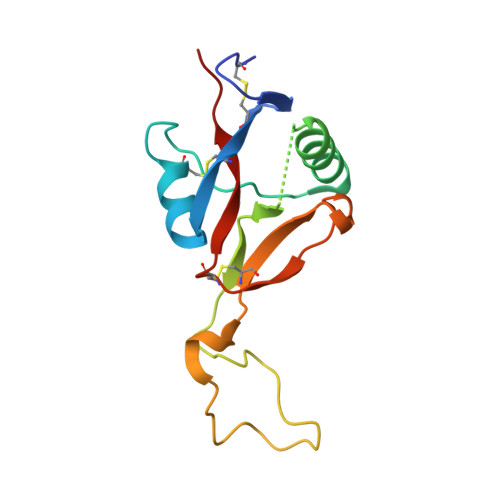
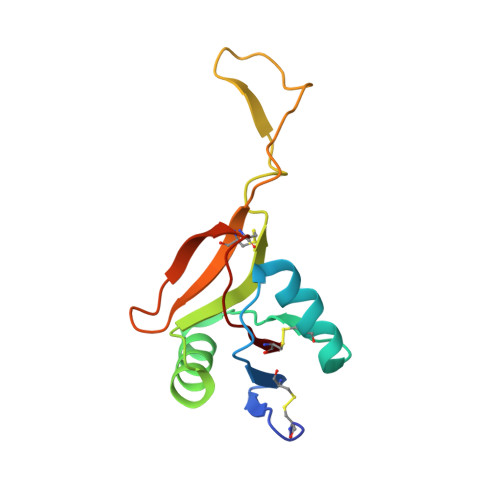
Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases.
Takeda, S., Igarashi, T., Mori, H.(2007) FEBS Lett 581: 5859-5864
- PubMed: 18060879
- DOI: https://doi.org/10.1016/j.febslet.2007.11.062
- Primary Citation of Related Structures:
2E3X - PubMed Abstract:
Russell's viper venom factor X activator (RVV-X) is a heterotrimeric metalloproteinase with a mammalian ADAM-like heavy chain and two lectin-like light chains. The crystal structure of RVV-X has been determined at 2.9 A resolution and shows a hook-spanner-wrench-like architecture, in which the metalloproteinase/disintegrin region constitutes a hook, and the lectin-like domains constitute a handle. A 6.5nm separation between the catalytic site and a putative exosite suggests a docking model for factor X. The structure provides a typical example of the molecular evolution of multi-subunit proteins and insights into the molecular basis of target recognition and proteolysis by ADAM/adamalysin/reprolysin proteinases.
- Department of Cardiac Physiology, National Cardiovascular Center Research Institute, 5-7-1 Fujishiro-dai, Suita, Osaka 565-8565, Japan. stakeda@ri.ncvc.go.jp
Organizational Affiliation: