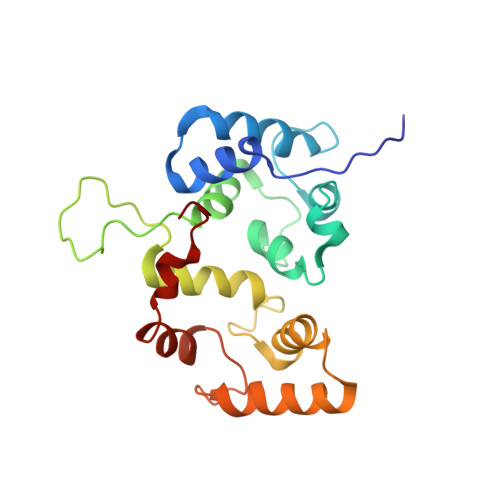
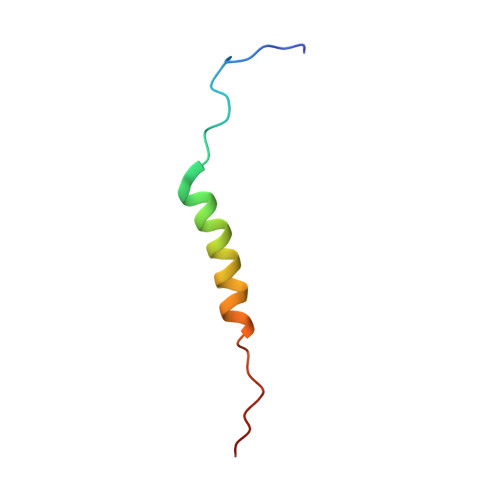
Solution structure of the cytoplasmic region of Na+/H+ exchanger 1 complexed with essential cofactor calcineurin B homologous protein 1
Mishima, M., Wakabayashi, S., Kojima, C.(2007) J Biological Chem 282: 2741-2751
- PubMed: 17050540
- DOI: https://doi.org/10.1074/jbc.M604092200
- Primary Citation of Related Structures:
2E30 - PubMed Abstract:
Na+/H+ exchanger 1 (NHE1) regulates intracellular pH, Na+ content, and cell volume. Calcineurin B homologous protein 1 (CHP1) serves as an essential cofactor that facilitates NHE1 exchange activity under physiological conditions by direct binding to the cytoplasmic juxtamembrane region of NHE1. Here we describe the solution structure of the cytoplasmic juxtamembrane region of NHE1 complexed with CHP1. The region of NHE1 forms an amphipathic helix, which is induced by CHP1 binding, and CHP1 possesses a large hydrophobic cleft formed by EF-hand helices. The apolar side of the NHE1 helix participates in extensive hydrophobic interactions with the cleft of CHP1. We suggest that helix formation of the cytoplasmic region of NHE1 by CHP1 is a prerequisite for generating the active form of NHE1. The molecular recognition detailed in this study also provides novel insight into the target binding mechanism of EF-hand proteins.
- Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: