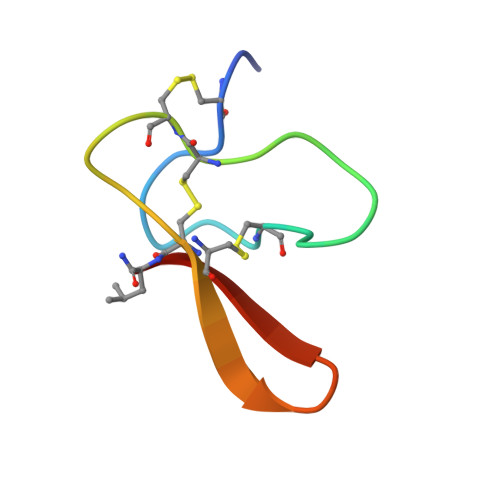
Solution structure of agelenin, an insecticidal peptide isolated from the spider Agelena opulenta, and its structural similarities to insect-specific calcium channel inhibitors
Yamaji, N., Sugase, K., Nakajima, T., Miki, T., Wakamori, M., Mori, Y., Iwashita, T.(2007) FEBS Lett 581: 3789-3794
- PubMed: 17644092
- DOI: https://doi.org/10.1016/j.febslet.2007.06.077
- Primary Citation of Related Structures:
2E2S - PubMed Abstract:
Agelenin, isolated from the Agelenidae spider Agelena opulenta, is a peptide composed of 35 amino acids. We determined the three-dimensional structure of agelenin using two-dimensional NMR spectroscopy. The structure is composed of a short antiparallel beta-sheet and four beta-turns, which are stabilized by three disulfide bonds. Agelenin has characteristic residues, Phe9, Ser28 and Arg33, which are arranged similarly to the pharmacophore of the insect channel inhibitor, omega-atracotoxin-Hv1a. These observations suggest that agelenin and omega-atracotoxin-Hv1a bind to insect calcium channels in a similar manner. We also suggest that another mode of action may operate in the channel inhibition by omega-agatoxin-IVA and omega-atracotoxin-Hv2a.
- Suntory Institute for Bioorganic Research, 1-1-1 Wakayamadai, Shimamoto-Cho, Mishima-Gun, Osaka 618-8503, Japan.
Organizational Affiliation: