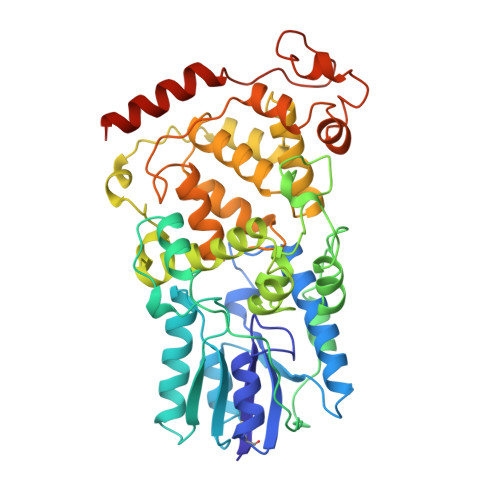
Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
Fujihashi, M., Numoto, N., Kobayashi, Y., Mizushima, A., Tsujimura, M., Nakamura, A., Kawarabayasi, Y., Miki, K.(2007) J Mol Biology 365: 903-910
- PubMed: 17107688
- DOI: https://doi.org/10.1016/j.jmb.2006.10.012
- Primary Citation of Related Structures:
2E0I - PubMed Abstract:
UV exposure of DNA molecules induces serious DNA lesions. The cyclobutane pyrimidine dimer (CPD) photolyase repairs CPD-type - lesions by using the energy of visible light. Two chromophores for different roles have been found in this enzyme family; one catalyzes the CPD repair reaction and the other works as an antenna pigment that harvests photon energy. The catalytic cofactor of all known photolyases is FAD, whereas several light-harvesting cofactors are found. Currently, 5,10-methenyltetrahydrofolate (MTHF), 8-hydroxy-5-deaza-riboflavin (8-HDF) and FMN are the known light-harvesting cofactors, and some photolyases lack the chromophore. Three crystal structures of photolyases from Escherichia coli (Ec-photolyase), Anacystis nidulans (An-photolyase), and Thermus thermophilus (Tt-photolyase) have been determined; however, no archaeal photolyase structure is available. A similarity search of archaeal genomic data indicated the presence of a homologous gene, ST0889, on Sulfolobus tokodaii strain7. An enzymatic assay reveals that ST0889 encodes photolyase from S. tokodaii (St-photolyase). We have determined the crystal structure of the St-photolyase protein to confirm its structural features and to investigate the mechanism of the archaeal DNA repair system with light energy. The crystal structure of the St-photolyase is superimposed very well on the three known photolyases including the catalytic cofactor FAD. Surprisingly, another FAD molecule is found at the position of the light-harvesting cofactor. This second FAD molecule is well accommodated in the crystal structure, suggesting that FAD works as a novel light-harvesting cofactor of photolyase. In addition, two of the four CPD recognition residues in the crystal structure of An-photolyase are not found in St-photolyase, which might utilize a different mechanism to recognize the CPD from that of An-photolyase.
- Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan.
Organizational Affiliation: