Structure of the Oncoprotein Gankyrin in Complex with S6 ATPase of the 26S Proteasome
Nakamura, Y., Nakano, K., Umehara, T., Tanaka, A., Padmanabhan, B., Yokoyama, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 26S proteasome non-ATPase regulatory subunit 10 | 231 | Mus musculus | Mutation(s): 0 | 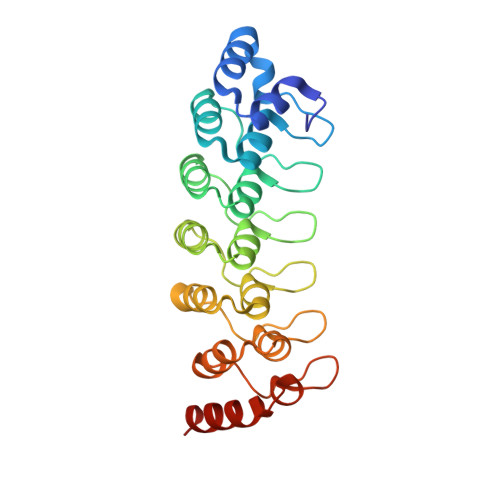 | |
UniProt | |||||
Find proteins for Q9Z2X2 (Mus musculus) Explore Q9Z2X2 Go to UniProtKB: Q9Z2X2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Z2X2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 26S protease regulatory subunit 6B | 83 | Rattus norvegicus | Mutation(s): 0 | 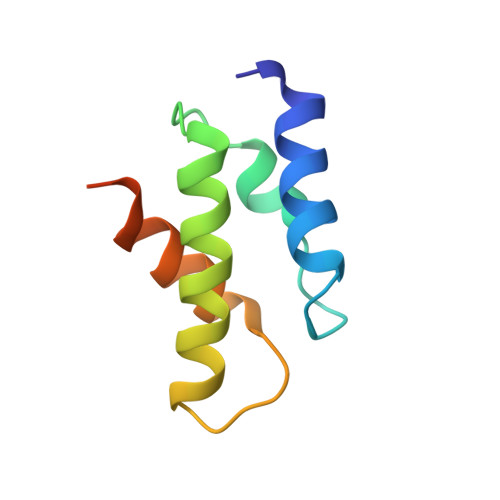 | |
UniProt | |||||
Find proteins for Q63570 (Rattus norvegicus) Explore Q63570 Go to UniProtKB: Q63570 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q63570 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | E [auth A] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 41.991 | α = 90 |
| b = 92.319 | β = 90 |
| c = 167.191 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOLREP | phasing |
| CNS | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |