Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
Li, S., Sandercock, A.M., Conduit, P., Robinson, C.V., Williams, R.L., Kilmartin, J.V.(2006) J Cell Biol 173: 867-877
- PubMed: 16785321
- DOI: https://doi.org/10.1083/jcb.200603153
- Primary Citation of Related Structures:
2DOQ, 2GV5 - PubMed Abstract:
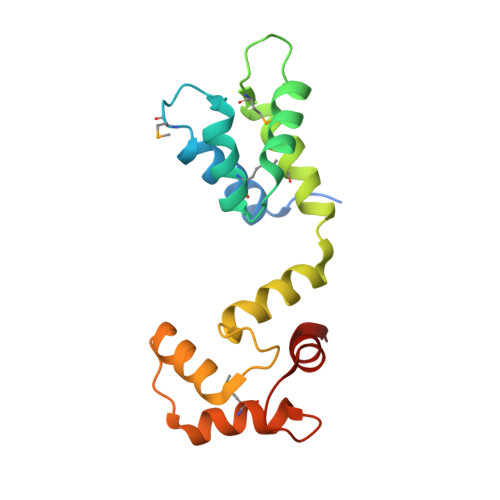
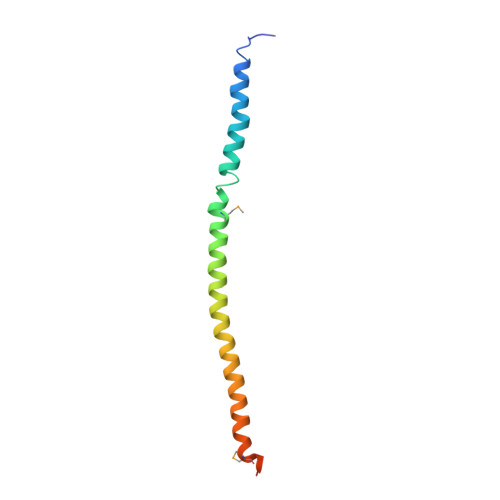
Centrins are calmodulin-like proteins present in centrosomes and yeast spindle pole bodies (SPBs) and have essential functions in their duplication. The Saccharomyces cerevisiae centrin, Cdc31p, binds Sfi1p on multiple conserved repeats; both proteins localize to the SPB half-bridge, where the new SPB is assembled. The crystal structures of Sfi1p-centrin complexes containing several repeats show Sfi1p as an alpha helix with centrins wrapped around each repeat and similar centrin-centrin contacts between each repeat. Electron microscopy (EM) shadowing of an Sfi1p-centrin complex with 15 Sfi1 repeats and 15 centrins bound showed filaments 60 nm long, compatible with all the Sfi1 repeats as a continuous alpha helix. Immuno-EM localization of the Sfi1p N and C termini showed Sfi1p-centrin filaments spanning the length of the half-bridge with the Sfi1p N terminus at the SPB. This suggests a model for SPB duplication where the half-bridge doubles in length by association of the Sfi1p C termini, thereby providing a new Sfi1p N terminus to initiate SPB assembly.
- Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 2QH, England, UK.
Organizational Affiliation: