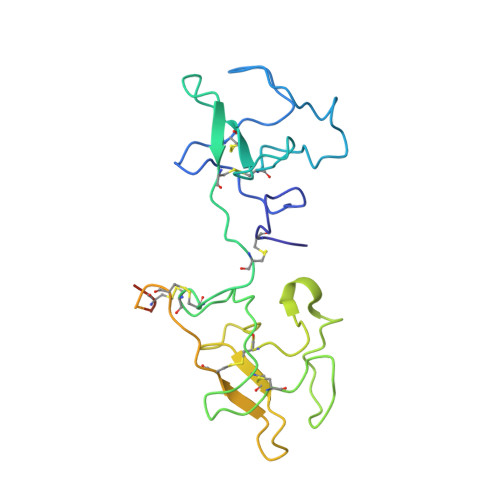
X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM
Cnudde, S.E., Prorok, M., Castellino, F.J., Geiger, J.H.(2006) Biochemistry 45: 11052-11060
- PubMed: 16964966
- DOI: https://doi.org/10.1021/bi060914j
- Primary Citation of Related Structures:
2DOH, 2DOI - PubMed Abstract:
The crystal structure of the human Pg-derived angiogenesis inhibitor, angiostatin, complexed to VEK-30, a peptide from the group A streptococcal surface protein, PAM, was determined and refined to 2.3 A resolution. This is the first structure of angiostatin bound to a ligand and provides a model of the interaction between Pg and streptococcal-derived pathogenic proteins. VEK-30 contains a "through-space isostere" for C-terminal lysine, wherein Arg and Glu side chains, separated by one helical turn, bind within the bipolar angiostatin kringle 2 (K2) domain lysine-binding site. VEK-30 also makes several contacts with K2 residues that exist outside of the canonical LBS and are not conserved among the other Pg kringles, thus providing a molecular basis for the selectivity of VEK-30 for K2. The structure also shows that Pg kringle domains undergo significant structural rearrangement relative to one another and reveals dimerization between two molecules of angiostatin and VEK-30 related by crystallographic symmetry. This dimerization, which exists only in the crystal structure, is consistent with the parallel coiled-coil full-length PAM dimer expected from sequence similarities and homology modeling.
- Department of Biochemistry, Michigan State University, East Lansing, Michigan 48824, USA.
Organizational Affiliation: