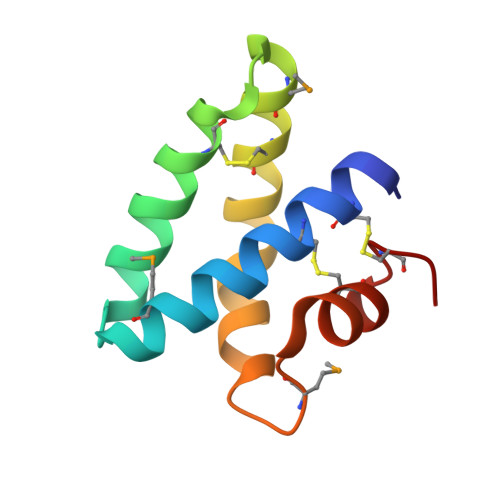
Crystal structures of saposins A and C.
Ahn, V.E., Leyko, P., Alattia, J.R., Chen, L., Prive, G.G.(2006) Protein Sci 15: 1849-1857
- PubMed: 16823039
- DOI: https://doi.org/10.1110/ps.062256606
- Primary Citation of Related Structures:
2DOB, 2GTG - PubMed Abstract:
Saposins A and C are sphingolipid activator proteins required for the lysosomal breakdown of galactosylceramide and glucosylceramide, respectively. The saposins interact with lipids, leading to an enhanced accessibility of the lipid headgroups to their cognate hydrolases. We have determined the crystal structures of human saposins A and C to 2.0 Angstroms and 2.4 Angstroms, respectively, and both reveal the compact, monomeric saposin fold. We confirmed that these two proteins were monomeric in solution at pH 7.0 by analytical centrifugation. However, at pH 4.8, in the presence of the detergent C(8)E(5), saposin A assembled into dimers, while saposin C formed trimers. Saposin B was dimeric under all conditions tested. The self-association of the saposins is likely to be relevant to how these small proteins interact with lipids, membranes, and hydrolase enzymes.
- Department of Medical Biophysics, University of Toronto, Canada.
Organizational Affiliation: