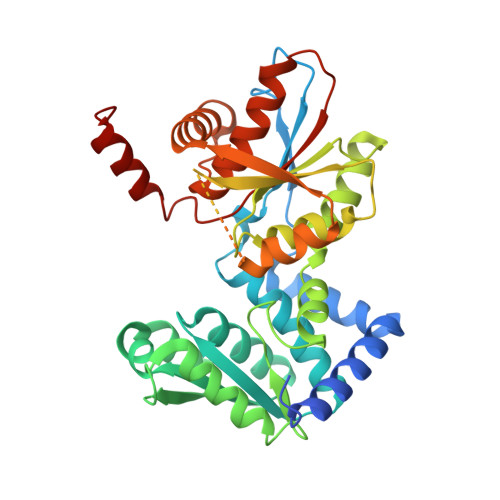
Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Nishio, K., Ogasahara, K., Morimoto, Y., Tsukihara, T., Lee, S.J., Yutani, K.(2010) FEBS J 277: 2157-2170
- PubMed: 20370823
- DOI: https://doi.org/10.1111/j.1742-4658.2010.07631.x
- Primary Citation of Related Structures:
2DH5, 2DH6 - PubMed Abstract:
To understand the basis for the lower activity of the tryptophan synthase beta(2) subunit in comparison to the alpha(2)beta(2) complex, we determined the crystal structures of apo-beta(2) and holo-beta(2) from Escherichia coli at 3.0 and 2.9 A resolutions, respectively. To our knowledge, this is the first report of both beta(2) subunit structures with and without pyridoxal-5'-phosphate. The apo-type molecule retained a dimeric form in solution, as in the case of the holo-beta(2) subunit. The subunit structures of both the apo-beta(2) and the holo-beta(2) forms consisted of two domains, namely the N domain and the C domain. Although there were significant structural differences between the apo- and holo-structures, they could be easily superimposed with a 22 degrees rigid body rotation of the C domain. The pyridoxal-5'-phosphate-bound holo-form had multiple interactions between the two domains and a long loop (residues 260-310), which were missing in the apo-form. Comparison of the structures of holo-Ecbeta(2) and Stbeta(2) in the alpha(2)beta(2) complex from Salmonella typhimurium (Stalpha(2)beta(2)) identified the cause of the lower enzymatic activity of holo-Ecbeta(2) in comparison with Stalpha(2)beta(2). The substrate (indole) gate residues, Tyr279 and Phe280, block entry of the substrate into the beta(2) subunit, although the indole can directly access the active site as a result of a wider cleft between the N and C domains in the holo-Ecbeta(2) subunit. In addition, the structure around betaAsp305 of the holo-Ecbeta(2) subunit was similar to the open state of Stalpha(2)beta(2) with low activity, resulting in lower activity of holo-Ecbeta(2).
- Institute for Protein Research, Osaka University, Japan.
Organizational Affiliation: