Bispecific abs against modified protein and DNA with oxidized lipids
Akagawa, M., Ito, S., Toyoda, K., Ishii, Y., Tatsuda, E., Shibata, T., Yamaguchi, S., Kawai, Y., Ishino, K., Kishi, Y., Adachi, T., Tsubata, T., Takasaki, Y., Hattori, N., Matsuda, T., Uchida, K.(2006) Proc Natl Acad Sci U S A 103: 6160-6165
- PubMed: 16603628
- DOI: https://doi.org/10.1073/pnas.0600865103
- Primary Citation of Related Structures:
2DDQ - PubMed Abstract:
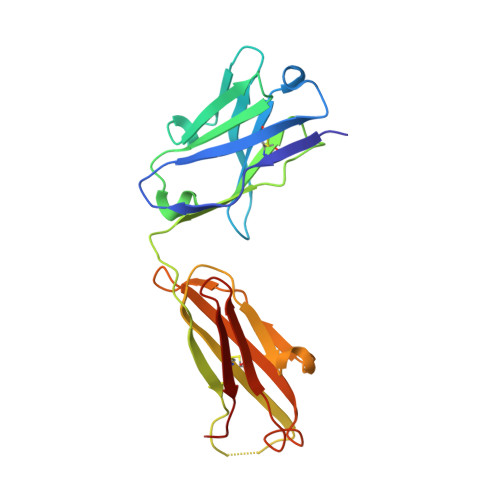
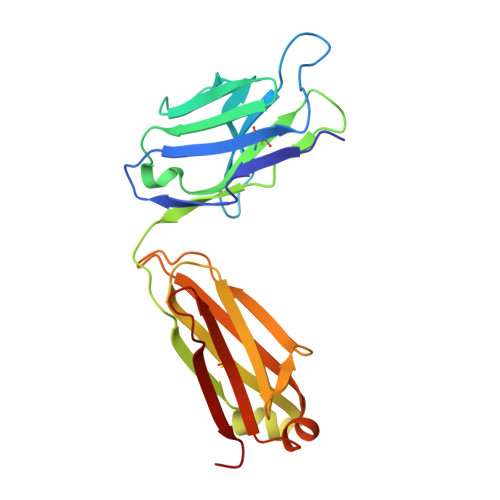
4-Hydroxy-2-nonenal (HNE), a racemic mixture of 4R- and 4S-enantiomers, is a major product of lipid peroxidation and is believed to be largely responsible for the cytopathological effects observed during oxidative stress. HNE reacts with histidine to form a stable HNE-histidine Michael addition-type adduct possessing three chiral centers in the cyclic hemiacetal structure. We have previously raised the mAbs, anti-R mAb 310 and anti-S mAb S412, that enantioselectively recognized the R-HNE-histidine and R-HNE-histidine adducts, respectively, and demonstrated the presence of both epitopes in vivo. In the present study, to further investigate the anti-HNE immune response, we analyzed the variable genes and primary structure of these Abs and found that the sequence of R310 was highly homologous to anti-DNA autoantibodies, the hallmark of systemic lupus erythematosus. An x-ray crystallographic analysis of the R310 Fab fragment showed that the R-HNE-histidine adduct binds to a hydrophobic pocket in the antigen-binding site. Despite the structural identity to the anti-DNA autoantibodies, however, R310 showed only a slight crossreactivity with the native double-stranded DNA, whereas the Ab immunoreactivity was dramatically enhanced by the treatment of the DNA with 4-oxo-2-nonenal (ONE), an analog of HNE. Moreover, the 7-(2-oxo-heptyl)-substituted 1,N2-etheno-type ONE-2'-deoxynucleoside adducts were identified as alternative epitopes of R310. Molecular mimicry between the R-HNE-histidine configurational isomers and the ONE-DNA base adducts is proposed for the dual crossreactivity.
- Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya 464-8601, Japan.
Organizational Affiliation: