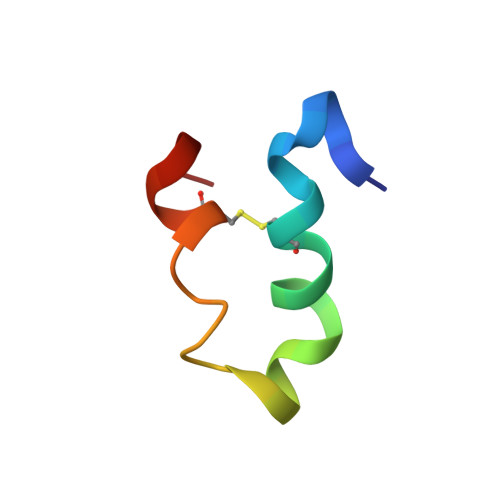
Peptide design and structural characterization of a GPCR loop mimetic
Pham, T.C.T., Kriwacki, R.W., Parrill, A.L.(2007) Biopolymers 86: 298-310
- PubMed: 17443712
- DOI: https://doi.org/10.1002/bip.20745
- Primary Citation of Related Structures:
2DCO - PubMed Abstract:
G protein-coupled receptors (GPCRs) control fundamental aspects of human physiology and behaviors. Knowledge of their structures, especially for the loop regions, is limited and has principally been obtained from homology models, mutagenesis data, low resolution structural studies, and high resolution studies of peptide models of receptor segments. We developed an alternate methodology for structurally characterizing GPCR loops, using the human S1P(4) first extracellular loop (E1) as a model system. This methodology uses computational peptide designs based on transmembrane domain (TM) model structures in combination with CD and NMR spectroscopy. The characterized peptides contain segments that mimic the self-assembling extracellular ends of TM 2 and TM 3 separated by E1, including residues R3.28(121) and E3.29(122) that are required for sphingosine 1-phosphate (S1P) binding and receptor activation in the S1P(4) receptor. The S1P(4) loop mimetic peptide interacted specifically with an S1P headgroup analog, O-phosphoethanolamine (PEA), as evidenced by PEA-induced perturbation of disulfide cross-linked coiled-coil first extracellular loop mimetic (CCE1a) (1)H and (15)N backbone amide chemical shifts. CCE1a was capable of weakly binding PEA near biologically relevant residues R29 and E30, which correspond to R3.28 and E3.29 in the full-length S1P(4) receptor, confirming that it has adopted a biologically relevant conformation. We propose that the combination of coiled-coil TM replacement and conformational stabilization with an interhelical disulfide bond is a general design strategy that promotes native-like structure for loops derived from GPCRs.
- Department of Chemistry and Computational Research on Materials Institute, The University of Memphis, Memphis, TN 38152, USA.
Organizational Affiliation: