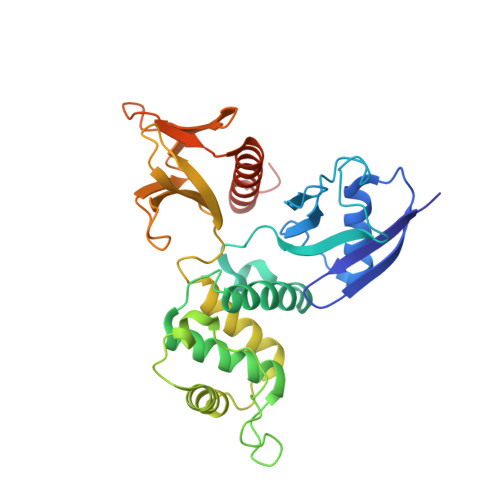
Structure of dimerized radixin FERM domain suggests a novel masking motif in C-terminal residues 295-304
Kitano, K., Yusa, F., Hakoshima, T.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 340-345
- PubMed: 16582480
- DOI: https://doi.org/10.1107/S1744309106010062
- Primary Citation of Related Structures:
2D2Q - PubMed Abstract:
ERM (ezrin/radixin/moesin) proteins bind to the cytoplasmic tail of adhesion molecules in the formation of the membrane-associated cytoskeleton. The binding site is located in the FERM (4.1 and ERM) domain, a domain that is masked in the inactive form. A conventional masking motif, strand 1 (residues 494-500 in radixin), has previously been identified in the C-terminal tail domain. Here, the crystal structure of dimerized radixin FERM domains (residues 1-310) is presented in which the binding site of one molecule is occupied by the C-terminal residues (residues 295-304, strand 2) of the other molecule. The residues contain a conserved motif that is compatible with that identified in the adhesion molecules. The residues might serve as a second masking region in the inactive form of ERM proteins.
- Structural Biology Laboratory, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: