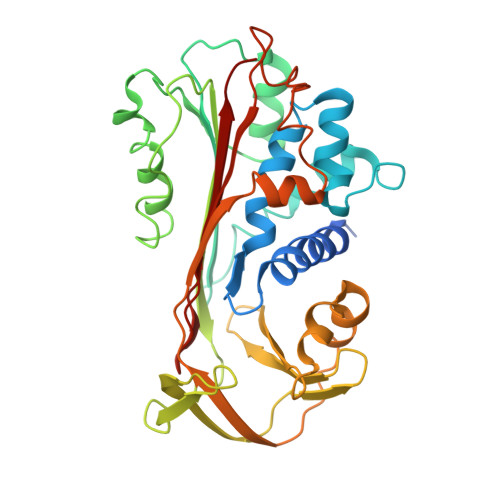
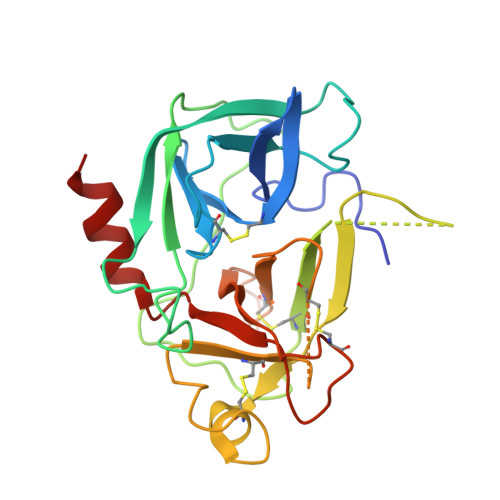
Active Site Distortion Is Sufficient for Proteinase Inhibition by Serpins: Structure of the covalent complex of alpha 1-proteinase inhibitor with porcine pancreatic elastase
Dementiev, A., Dobo, J., Gettins, P.G.(2006) J Biological Chem 281: 3452-3457
- PubMed: 16321984
- DOI: https://doi.org/10.1074/jbc.M510564200
- Primary Citation of Related Structures:
2D26 - PubMed Abstract:
We report here the x-ray structure of a covalent serpin-proteinase complex, alpha1-proteinase inhibitor (alpha1PI) with porcine pancreatic elastase (PPE), which differs from the only other x-ray structure of such a complex, that of alpha1PI with trypsin, in showing nearly complete definition of the proteinase. alpha1PI complexes with trypsin, PPE, and human neutrophil elastase (HNE) showed similar rates of deacylation and enhanced susceptibility to proteolysis by exogenous proteinases in solution. The differences between the two x-ray structures therefore cannot arise from intrinsic differences in the inhibition mechanism. However, self-proteolysis of purified complex resulted in rapid cleavage of the trypsin complex, slower cleavage of the PPE complex, and only minimal cleavage of the HNE complex. This suggests that the earlier alpha1 PI-trypsin complex may have been proteolyzed and that the present structure is more likely to be representative of serpin-proteinase complexes. The present structure shows that active site distortion alone is sufficient for inhibition and suggests that enhanced proteolysis is not necessarily exploited in vivo.
- Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, Illinois 60607, USA.
Organizational Affiliation: