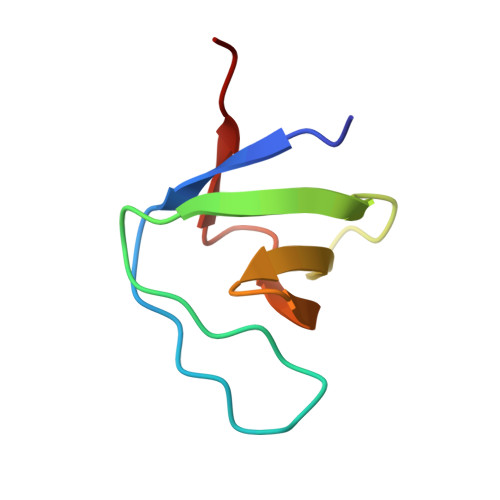
Crystal structure of the C-terminal SH3 domain of the adaptor protein GADS in complex with SLP-76 motif peptide reveals a unique SH3-SH3 interaction
Dimasi, N.(2007) Int J Biochem Cell Biol 39: 109-123
- PubMed: 17010654
- DOI: https://doi.org/10.1016/j.biocel.2006.07.003
- Primary Citation of Related Structures:
2D0N - PubMed Abstract:
The Grb2-like adaptor protein GADS is essential for tyrosine kinase-dependent signaling in T lymphocytes. Following T cell receptor ligation, GADS interacts through its C-terminal SH3 domain with the adaptors SLP-76 and LAT, to form a multiprotein signaling complex that is crucial for T cell activation. To understand the structural basis for the selective recognition of GADS by SLP-76, herein is reported the crystal structure at 1.54 Angstrom of the C-terminal SH3 domain of GADS bound to the SLP-76 motif 233-PSIDRSTKP-241, which represents the minimal binding site. In addition to the unique structural features adopted by the bound SLP-76 peptide, the complex structure reveals a unique SH3-SH3 interaction. This homophilic interaction, which is observed in presence of the SLP-76 peptide and is present in solution, extends our understanding of the molecular mechanisms that could be employed by modular proteins to increase their signaling transduction specificity.
- Laboratory for Molecular Medicine, Istituto Giannina Gaslini, Largo Gerolamo Gaslini 5, Genova 16147, Italy. ndimasi@gmail.com
Organizational Affiliation: