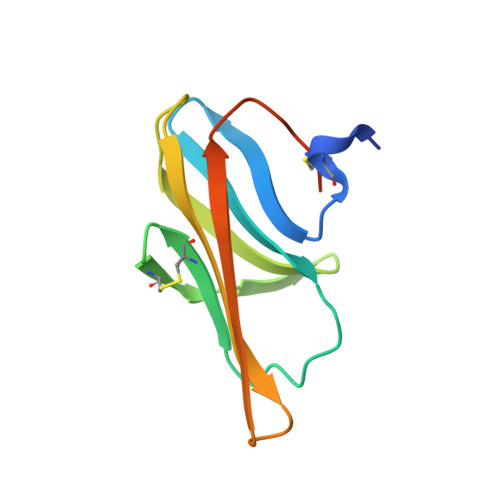
The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1,3-xylan
Hashimoto, H., Tamai, Y., Okazaki, F., Tamaru, Y., Shimizu, T., Araki, T., Sato, M.(2005) FEBS Lett 579: 4324-4328
- PubMed: 16061225
- DOI: https://doi.org/10.1016/j.febslet.2005.06.062
- Primary Citation of Related Structures:
2COV - PubMed Abstract:
Here, we present the crystal structure of the family 31 carbohydrate-binding module (CBM) of beta-1,3-xylanase from Alcaligenes sp. strain XY-234 (AlcCBM31) determined at a resolution of 1.25A. The AlcCBM31 shows affinity with only beta-1,3-xylan. The AlcCBM31 molecule makes a beta-sandwich structure composed of eight beta-strands with a typical immunoglobulin fold and contains two intra-molecular disulfide bonds. The folding topology of AlcCBM31 differs from that of the large majority of other CBMs, in which eight beta-strands comprise a beta-sandwich structure with a typical jelly-roll fold. AlcCBM31 shows structural similarity with CBM structures of family 34 and family 9, which also adopt structures based on immunoglobulin folds.
- International Graduate school of Arts and Sciences, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan. hash@tsurumi.yokohama-cu.ac.jp
Organizational Affiliation: