Structural Basis for HIV-1 Neutralization by a Gp41 Fusion Intermediate-Directed Antibody
Luftig, M.A., Mattu, M., Di Giovine, P., Geleziunas, R., Hrin, R., Barbato, G., Bianchi, E., Miller, M.D., Pessi, A., Carfi, A.(2006) Nat Struct Mol Biol 13: 740
- PubMed: 16862157
- DOI: https://doi.org/10.1038/nsmb1127
- Primary Citation of Related Structures:
2CMR - PubMed Abstract:
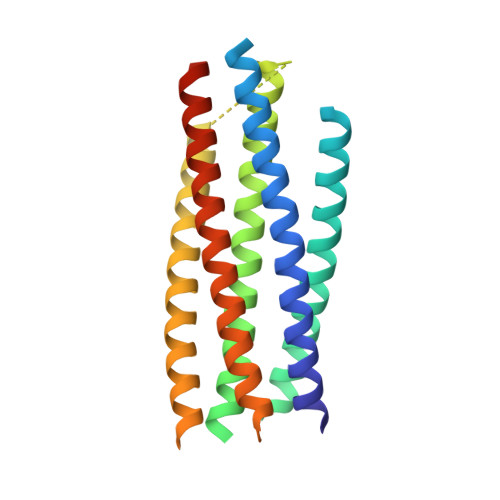
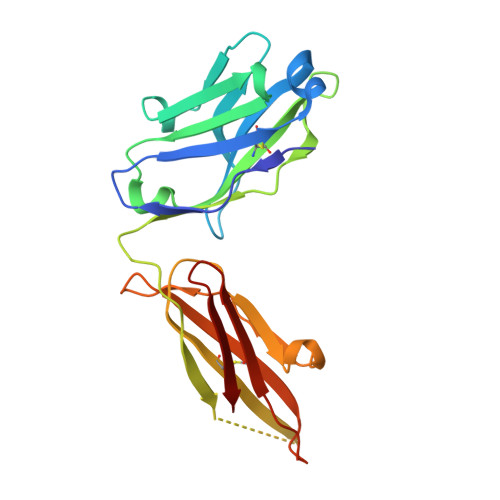
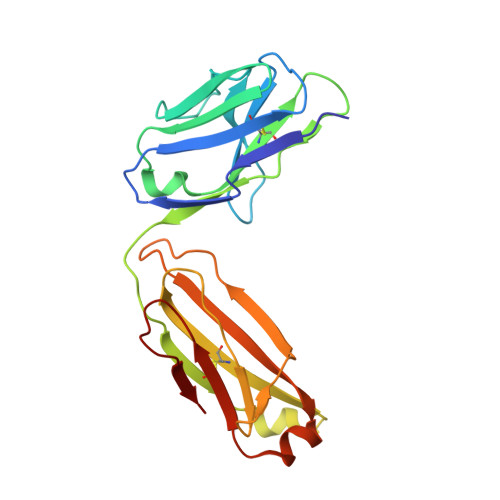
Elicitation of potent and broadly neutralizing antibodies is an important goal in designing an effective human immunodeficiency virus-1 (HIV-1) vaccine. The HIV-1 gp41 inner-core trimer represents a functionally and structurally conserved target for therapeutics. Here we report the 2.0-A-resolution crystal structure of the complex between the antigen-binding fragment of D5, an HIV-1 cross-neutralizing antibody, and 5-helix, a gp41 inner-core mimetic. Both binding and neutralization depend on residues in the D5 CDR H2 loop protruding into the conserved gp41 hydrophobic pocket, as well as a large pocket in D5 surrounding core gp41 residues. Kinetic analysis of D5 mutants with perturbed D5-gp41 interactions suggests that D5 persistence at the fusion intermediate is crucial for neutralization. Thus, our data validate the gp41 N-peptide trimer fusion intermediate as a target for neutralizing antibodies and provide a template for identification of more potent and broadly neutralizing molecules.
- Istituto di Ricerche di Biologia Molecolare P. Angeletti, Via Pontina Km 30,600, I-00040 Pomezia (Rome), Italy.
Organizational Affiliation: