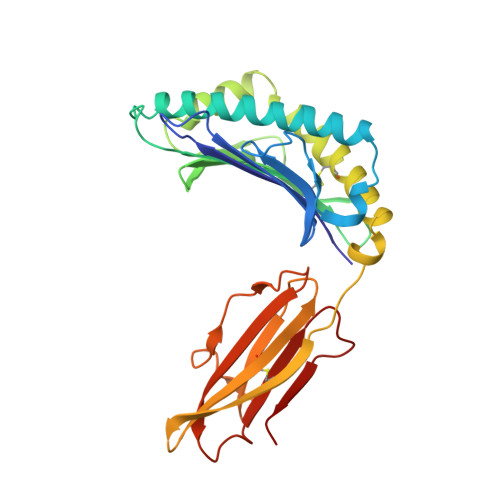
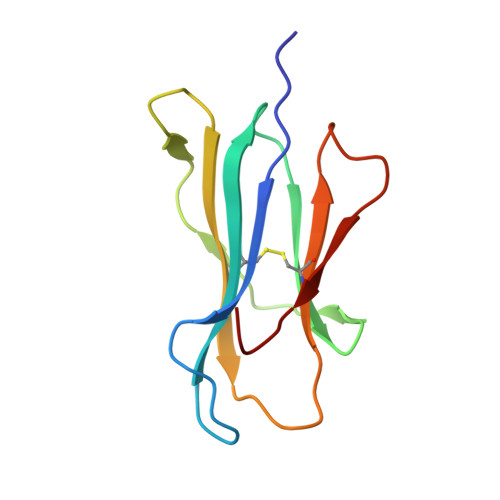
Three-dimensional structure of a peptide extending from one end of a class I MHC binding site.
Collins, E.J., Garboczi, D.N., Wiley, D.C.(1994) Nature 371: 626-629
- PubMed: 7935798
- DOI: https://doi.org/10.1038/371626a0
- Primary Citation of Related Structures:
2CLR - PubMed Abstract:
Class I major histocompatibility complex (MHC) molecules present peptides to CD8+ T cells for immunological surveillance (reviewed in ref. 1). The structures of complexes of class I MHC molecules with octamer, nonamer and decamer peptides determined until now show a common binding mode, with both peptide termini bound in conserved pockets at the ends of the peptide binding site. Length variations were accommodated by the peptide bulging or zig-zagging in the middle. Here we describe the structure of a decamer peptide which binds with the carboxy-terminal residue positioned outside the peptide binding site. Several protein side chains have rearranged to allow the peptide to exit. The structure suggests that even longer peptides could bind. The energetic effect of the altered mode of binding has been assessed by measuring the stability of the complex to thermal denaturation. Peptides bound in this novel manner are stable at physiological temperature, raising questions about their role in T-cell recognition and their production by proteolytic processing.
- Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts 02138.
Organizational Affiliation: