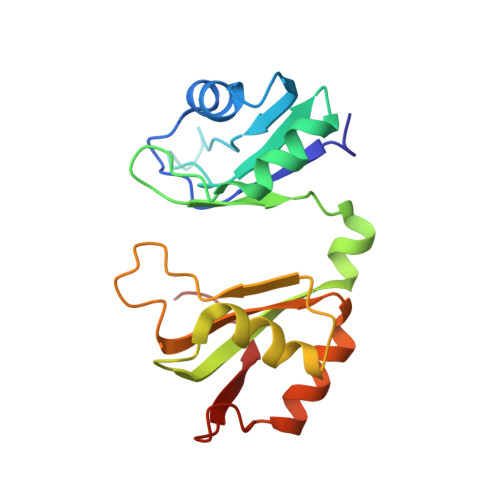
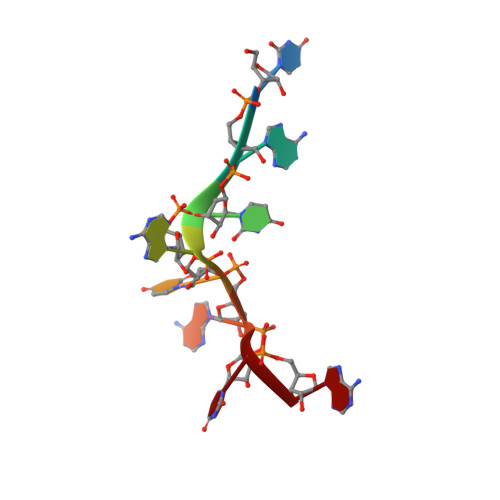
Grabbing the message: structural basis of mRNA 3'UTR recognition by Hrp1.
Perez-Canadillas, J.M.(2006) EMBO J 25: 3167-3178
- PubMed: 16794580
- DOI: https://doi.org/10.1038/sj.emboj.7601190
- Primary Citation of Related Structures:
2CJK - PubMed Abstract:
The recognition of specific signals encoded within the 3'-untranslated region of the newly transcribed mRNA triggers the assembly of a multiprotein machine that modifies its 3'-end. Hrp1 recognises one of such signals, the so-called polyadenylation enhancement element (PEE), promoting the recruitment of other polyadenylation factors in yeast. The molecular bases of this interaction are revealed here by the solution structure of a complex between Hrp1 and an oligonucleotide mimicking the PEE. Six consecutive bases (AUAUAU) are specifically recognised by two RNA-binding domains arranged in tandem. Both protein and RNA undergo significant conformational changes upon complex formation with a concomitant large surface burial of RNA bases. Key aspects of RNA specificity can be explained by the presence of intermolecular aromatic-aromatic contacts and hydrogen bonds. Altogether, the Hrp1-PEE structure represents one of the first steps towards understanding of the assembly of the cleavage and polyadenylation machinery at the atomic level.
- Laboratory of Molecular Biology, Medical Research Council, Cambridge, UK. jmperez@iqfr.csic.es
Organizational Affiliation: