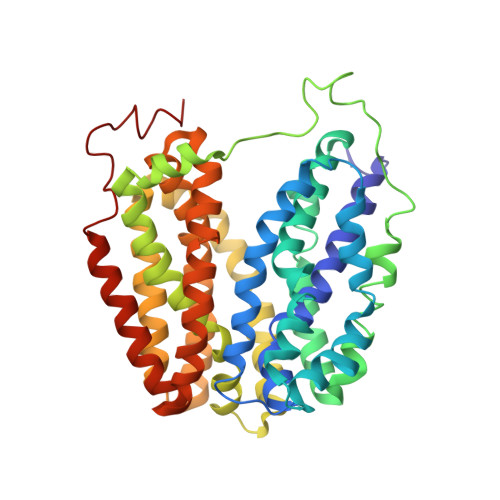
Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Mirza, O., Guan, L., Verner, G., Iwata, S., Kaback, H.R.(2006) EMBO J 25: 2038
- PubMed: 16525509
- DOI: https://doi.org/10.1038/sj.emboj.7601028
- Primary Citation of Related Structures:
2CFP, 2CFQ - PubMed Abstract:
Cation-coupled active transport is an essential cellular process found ubiquitously in all living organisms. Here, we present two novel ligand-free X-ray structures of the lactose permease (LacY) of Escherichia coli determined at acidic and neutral pH, and propose a model for the mechanism of coupling between lactose and H+ translocation. No sugar-binding site is observed in the absence of ligand, and deprotonation of the key residue Glu269 is associated with ligand binding. Thus, substrate induces formation of the sugar-binding site, as well as the initial step in H+ transduction.
- Department of Biological Sciences, Membrane Protein Crystallography Group, Imperial College London, London, UK.
Organizational Affiliation: