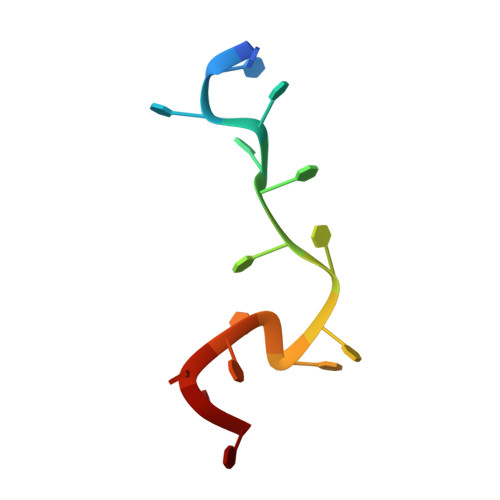Complexed Crystal Structure of Replication Restart Primsome Protein Prib Reveals a Novel Single-Stranded DNA-Binding Mode.
Huang, C.-Y., Hsu, C.-H., Sun, Y.-J., Wu, H.-N., Hsiao, C.-D.(2006) Nucleic Acids Res 34: 3878
- PubMed: 16899446
- DOI: https://doi.org/10.1093/nar/gkl536
- Primary Citation of Related Structures:
2CCZ - PubMed Abstract:
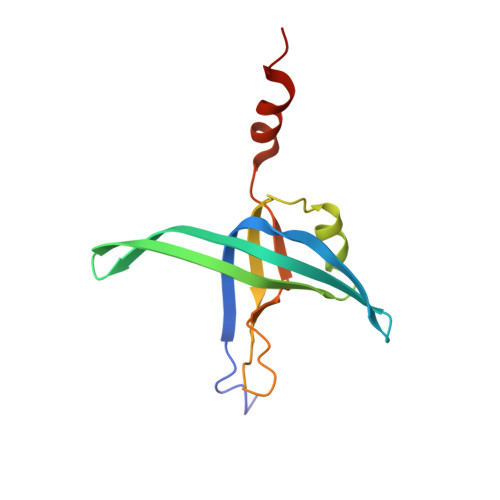
PriB is a primosomal protein required for replication restart in Escherichia coli. PriB stimulates PriA helicase activity via interaction with single-stranded DNA (ssDNA), but the molecular details of this interaction remain unclear. Here, we report the crystal structure of PriB complexed with a 15 bases oligonucleotide (dT15) at 2.7 A resolution. PriB shares structural similarity with the E.coli ssDNA-binding protein (EcoSSB). However, the structure of the PriB-dT15 complex reveals that PriB binds ssDNA differently. Results from filter-binding assays show that PriB-ssDNA interaction is salt-sensitive and cooperative. Mutational analysis suggests that the loop L45 plays an important role in ssDNA binding. Based on the crystal structure and biochemical analyses, we propose a cooperative mechanism for the binding of PriB to ssDNA and a model for the assembly of the PriA-PriB-ssDNA complex. This report presents the first structure of a replication restart primosomal protein complexed with DNA, and a novel model that explains the interactions between a dimeric oligonucleotide-binding-fold protein and ssDNA.
- Institute of Molecular Biology, Academia Sinica, Taipei, 115, Taiwan.
Organizational Affiliation: