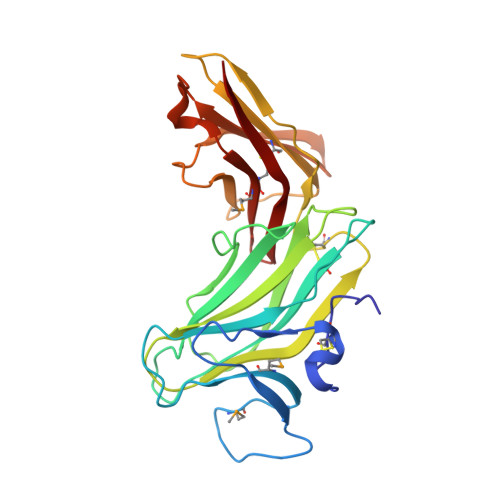
Molecular Analysis of Receptor Protein Tyrosine Phosphatase Mu-Mediated Cell Adhesion.
Aricescu, A.R., Hon, W.C., Siebold, C., Lu, W., Van Der Merwe, P.A., Jones, E.Y.(2006) EMBO J 25: 701
- PubMed: 16456543
- DOI: https://doi.org/10.1038/sj.emboj.7600974
- Primary Citation of Related Structures:
2C9A - PubMed Abstract:
Type IIB receptor protein tyrosine phosphatases (RPTPs) are bi-functional cell surface molecules. Their ectodomains mediate stable, homophilic, cell-adhesive interactions, whereas the intracellular catalytic regions can modulate the phosphorylation state of cadherin/catenin complexes. We describe a systematic investigation of the cell-adhesive properties of the extracellular region of RPTPmu, a prototypical type IIB RPTP. The crystal structure of a construct comprising its N-terminal MAM (meprin/A5/mu) and Ig domains was determined at 2.7 A resolution; this assigns the MAM fold to the jelly-roll family and reveals extensive interactions between the two domains, which form a rigid structural unit. Structure-based site-directed mutagenesis, serial domain deletions and cell-adhesion assays allowed us to identify the four N-terminal domains (MAM, Ig, fibronectin type III (FNIII)-1 and FNIII-2) as a minimal functional unit. Biophysical characterization revealed at least two independent types of homophilic interaction which, taken together, suggest that there is the potential for formation of a complex and possibly ordered array of receptor molecules at cell contact sites.
- Division of Structural Biology, Henry Wellcome Building of Genomic Medicine, University of Oxford, Oxford, UK.
Organizational Affiliation: