Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
Contreras-Martel, C., Job, V., Di Guilmi, A.-M., Vernet, T., Dideberg, O., Dessen, A.(2006) J Mol Biology 355: 684
- PubMed: 16316661
- DOI: https://doi.org/10.1016/j.jmb.2005.10.030
- Primary Citation of Related Structures:
2C5W, 2C6W - PubMed Abstract:
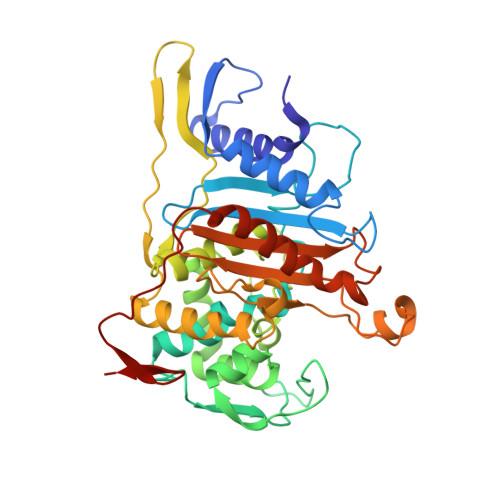
Streptococcus pneumoniae is a major human pathogen whose infections have been treated with beta-lactam antibiotics for over 60 years, but the proliferation of strains that are highly resistant to such drugs is a problem of worldwide concern. Beta-lactams target penicillin-binding proteins (PBPs), membrane-associated enzymes that play essential roles in the peptidoglycan biosynthetic process. Bifunctional PBPs catalyze both the polymerization of glycan chains (glycosyltransfer) and the cross-linking of adjacent pentapeptides (transpeptidation), while monofunctional enzymes catalyze only the latter reaction. Although S. pneumoniae has six PBPs, only three (PBP1a, PBP2x, PBP2b) are major resistance determinants, with PBP1a being the only bifunctional enzyme. PBP1a plays a key role in septum formation during the cell division cycle and its modification is essential for the development of high-level resistance to penicillins and cephalosporins. The crystal structure of a soluble form of pneumococcal PBP1a (PBP1a*) has been solved to 2.6A and reveals that it folds into three domains. The N terminus contains a peptide from the glycosyltransfer domain bound to an interdomain linker region, followed by a central, transpeptidase domain, and a small C-terminal unit. An analysis of PBP1a sequences from drug-resistant clinical strains in light of the structure reveals the existence of a mutational hotspot at the entrance of the catalytic cleft that leads to the modification of the polarity and accessibility of the mutated PBP1a active site. The presence of this hotspot in all variants sequenced to date is of key relevance for the development of novel antibiotherapies for the treatment of beta-lactam-resistant pneumococcal strains.
- Institut de Biologie Structurale Jean-Pierre Ebel (CNRS/CEA/UJF), Laboratoire de Cristallographie Macromoléculaire, 41 rue Jules Horowitz, Grenoble 38027, France.
Organizational Affiliation: