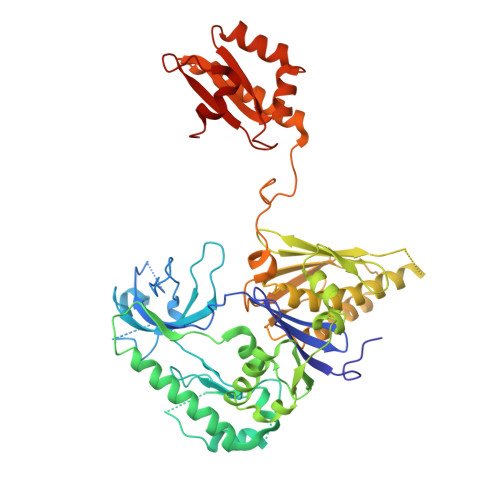
Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Callaghan, A.J., Marcaida, M.J., Stead, J.A., Mcdowall, K.J., Scott, W.G., Luisi, B.F.(2005) Nature 437: 1187
- PubMed: 16237448
- DOI: https://doi.org/10.1038/nature04084
- Primary Citation of Related Structures:
2BX2, 2C0B, 2C4R - PubMed Abstract:
The coordinated regulation of gene expression is required for homeostasis, growth and development in all organisms. Such coordination may be partly achieved at the level of messenger RNA stability, in which the targeted destruction of subsets of transcripts generates the potential for cross-regulating metabolic pathways. In Escherichia coli, the balance and composition of the transcript population is affected by RNase E, an essential endoribonuclease that not only turns over RNA but also processes certain key RNA precursors. RNase E cleaves RNA internally, but its catalytic power is determined by the 5' terminus of the substrate, even if this lies at a distance from the cutting site. Here we report crystal structures of the catalytic domain of RNase E as trapped allosteric intermediates with RNA substrates. Four subunits of RNase E catalytic domain associate into an interwoven quaternary structure, explaining why the subunit organization is required for catalytic activity. The subdomain encompassing the active site is structurally congruent to a deoxyribonuclease, making an unexpected link in the evolutionary history of RNA and DNA nucleases. The structure explains how the recognition of the 5' terminus of the substrate may trigger catalysis and also sheds light on the question of how RNase E might selectively process, rather than destroy, specific RNA precursors.
- Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK.
Organizational Affiliation: