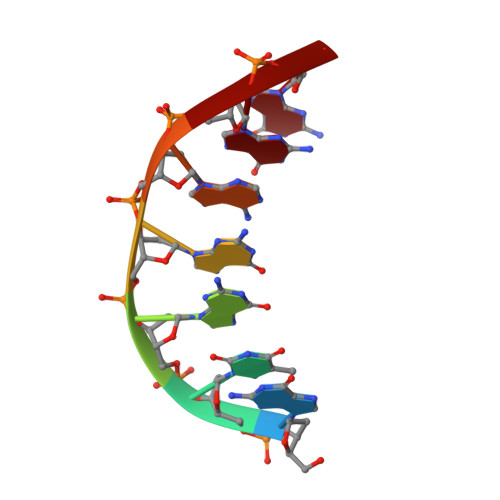
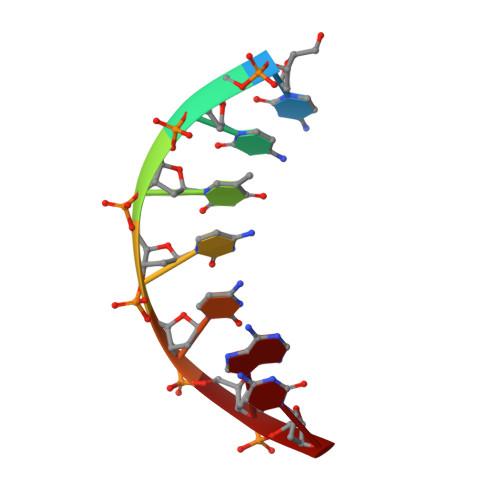
Structural Basis for DNA Bridging by Barrier-to-Autointegration Factor.
Bradley, C.M., Ronning, D.R., Ghirlando, R., Craigie, R., Dyda, F.(2005) Nat Struct Mol Biol 12: 935
- PubMed: 16155580
- DOI: https://doi.org/10.1038/nsmb989
- Primary Citation of Related Structures:
2BZF - PubMed Abstract:
The ability of barrier-to-autointegration factor (BAF) to bind and bridge DNA in a sequence-independent manner is crucial for its role in retroviral integration and a variety of cellular processes. To better understand this behavior, we solved the crystal structure of BAF bound to DNA. The structure reveals that BAF bridges DNA using two pairs of helix-hairpin-helix motifs located on opposite surfaces of the BAF dimer without changing its conformation.
- Laboratory of Molecular Biology, National Institute of Diabetes & Digestive & Kidney Diseases, US National Institutes of Health (NIH), 5 Center Drive, Bethesda, Maryland 20892, USA.
Organizational Affiliation: