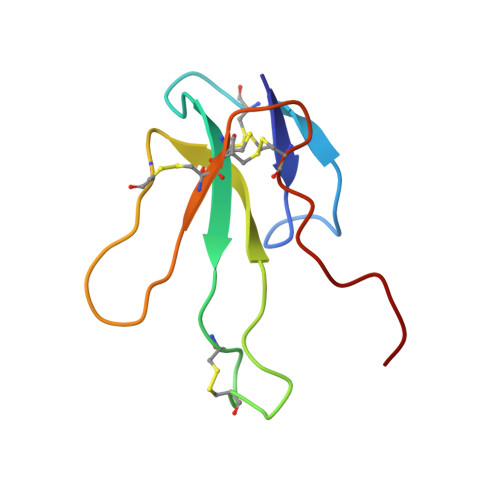
Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Scherf, T., Balass, M., Fuchs, S., Katchalski-Katzir, E., Anglister, J.(1997) Proc Natl Acad Sci U S A 94: 6059-6064
- PubMed: 9177168
- DOI: https://doi.org/10.1073/pnas.94.12.6059
- Primary Citation of Related Structures:
1BXP, 2BTX - PubMed Abstract:
The solution structure of the complex between alpha-bungarotoxin (alpha-BTX) and a 13-residue library-derived peptide (MRYYESSLKSYPD) has been solved using two-dimensional proton-NMR spectroscopy. The bound peptide adopts an almost-globular conformation resulting from three turns that surround a hydrophobic core formed by Tyr-11 of the peptide. The peptide fills an alpha-BTX pocket made of residues located at fingers I and II, as well as at the C-terminal region. Of the peptide residues, the largest contact area is formed by Tyr-3 and Tyr-4. These findings are in accord with the previous data in which it had been shown that substitution of these aromatic residues by aliphatic amino acids leads to loss of binding of the modified peptide with alpha-BTX. Glu-5 and Leu-8, which also remarkably contribute to the contact area with the toxin, are present in all the library-derived peptides that bind strongly to alpha-BTX. The structure of the complex may explain the fact that the library-derived peptide binds alpha-BTX with a 15-fold higher affinity than that shown by the acetylcholine receptor peptide (alpha185-196). Although both peptides bind to similar sites on alpha-BTX, the latter adopts an extended conformation when bound to the toxin [Basus, V., Song, G. & Hawrot, E. (1993) Biochemistry 32, 12290-12298], whereas the library peptide is nearly globular and occupies a larger surface area of alpha-BTX binding site.
- Department of Structural Biology, The Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: