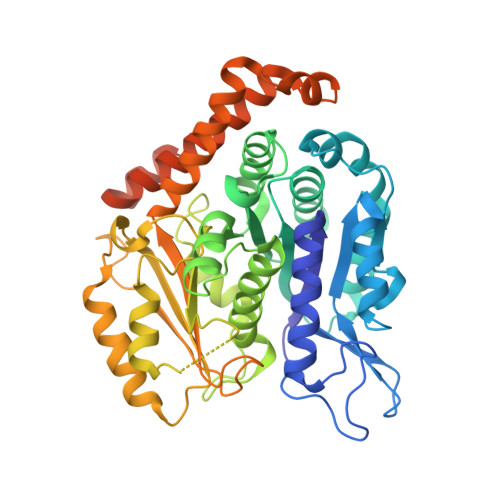
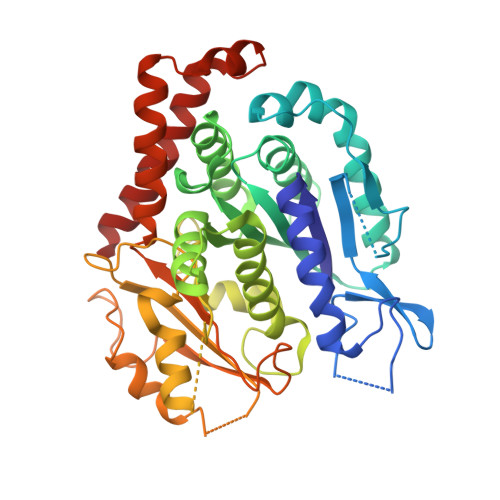
Structure of Bacterial Tubulin Btuba/B: Evidence for Horizontal Gene Transfer.
Schlieper, D., Oliva, M.A., Andreu, J.M., Lowe, J.(2005) Proc Natl Acad Sci U S A 102: 9170
- PubMed: 15967998
- DOI: https://doi.org/10.1073/pnas.0502859102
- Primary Citation of Related Structures:
2BTO, 2BTQ - PubMed Abstract:
alphabeta-Tubulin heterodimers, from which the microtubules of the cytoskeleton are built, have a complex chaperone-dependent folding pathway. They are thought to be unique to eukaryotes, whereas the homologue FtsZ can be found in bacteria. The exceptions are BtubA and BtubB from Prosthecobacter, which have higher sequence homology to eukaryotic tubulin than to FtsZ. Here we show that some of their properties are different from tubulin, such as weak dimerization and chaperone-independent folding. However, their structure is strikingly similar to tubulin including surface loops, and BtubA/B form tubulin-like protofilaments. Presumably, BtubA/B were transferred from a eukaryotic cell by horizontal gene transfer because their high degree of similarity to eukaryotic genes is unique within the Prosthecobacter genome.
- Laboratory of Molecular Biology, Medical Research Council, Hills Road, Cambridge CB2 2QH, United Kingdom.
Organizational Affiliation: