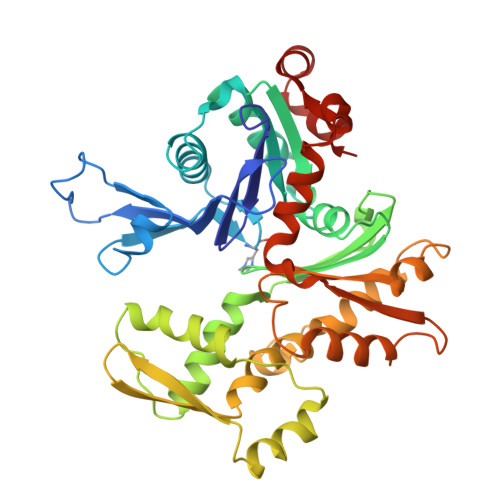
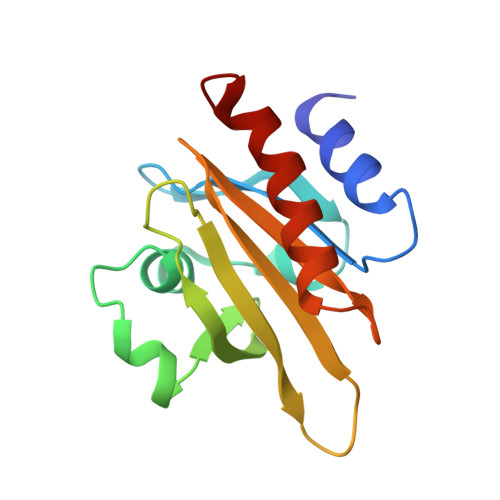
The structure of crystalline profilin-beta-actin.
Schutt, C.E., Myslik, J.C., Rozycki, M.D., Goonesekere, N.C., Lindberg, U.(1993) Nature 365: 810-816
- PubMed: 8413665
- DOI: https://doi.org/10.1038/365810a0
- Primary Citation of Related Structures:
2BTF - PubMed Abstract:
The three-dimensional structure of bovine profilin-beta-actin has been solved to 2.55 A resolution by X-ray crystallography. There are several significant local changes in the structure of beta-actin compared with alpha-actin as well as an overall 5 degrees rotation between its two major domains. Actin molecules in the crystal are organized into ribbons through intermolecular contacts like those found in oligomeric protein assemblies. Profilin forms two extensive contacts with the actin ribbon, one of which appears to correspond to the solution contact in vitro.
- Department of Chemistry, Henry H. Hoyt Laboratory, Princeton University, New Jersey 08544.
Organizational Affiliation: