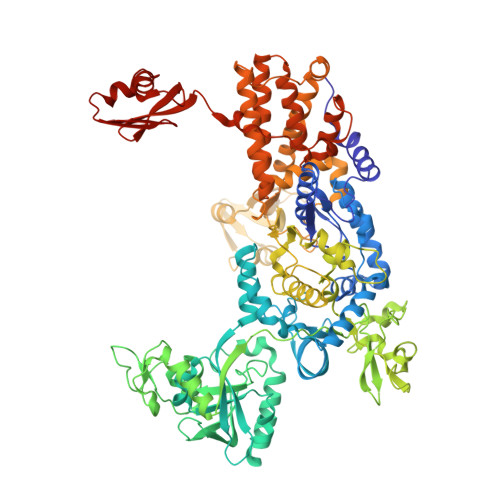
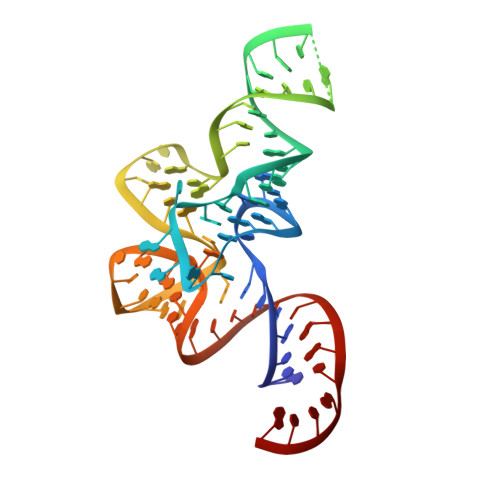
The Crystal Structure of Leucyl-tRNA Synthetase Complexed with tRNA(Leu) in the Post-Transfer- Editing Conformation.
Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S., Cusack, S.(2005) Nat Struct Mol Biol 12: 923
- PubMed: 16155583
- DOI: https://doi.org/10.1038/nsmb986
- Primary Citation of Related Structures:
2BTE, 2BYT - PubMed Abstract:
Leucyl-tRNA synthetase (LeuRS) has a specific post-transfer editing activity directed against mischarged isoleucine and similar noncognate amino acids. We describe the post-transfer-editing and product complexes of Thermus thermophilus LeuRS (LeuRSTT) with tRNA(Leu) at 2.9- to 3.3-A resolution. In the post-transfer-editing configuration, A76 binds in the editing active site exactly as previously found for the adenosine moiety of a small-molecule editing-substrate analog. The 60 C-terminal residues of LeuRSTT, unseen in previous structures, fold into a compact domain flexibly linked to the rest of the molecule and interacting with the G19-C56 tertiary base pair of tRNA(Leu). LeuRS recognition of tRNA(Leu) depends essentially on tRNA shape rather than base-specific interactions. The structures show that considerable domain rotations, notably of the editing domain, accompany the tRNA-3' end dynamics associated successively with aminoacylation, post-transfer editing and product release.
- European Molecular Biology Laboratory, Grenoble Outstation, c/o Institut Laue-Langevin, 156X, F-38042 Grenoble Cedex 9, France.
Organizational Affiliation: