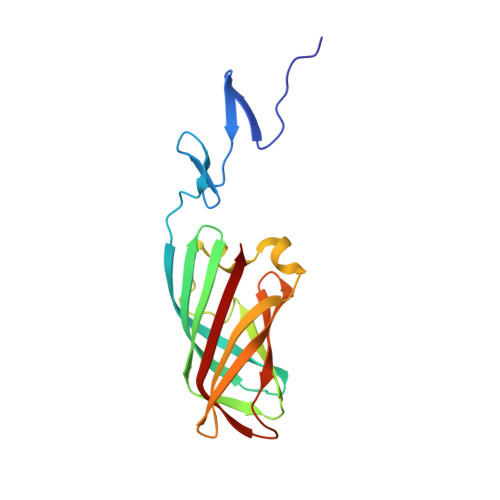
Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
Guardado Calvo, P., Fox, G.C., Hermo Parrado, X.L., Llamas-Saiz, A.L., Costas, C., Martinez-Costas, J., Benavente, J., van Raaij, M.J.(2005) J Mol Biology 354: 137
- PubMed: 16236316
- DOI: https://doi.org/10.1016/j.jmb.2005.09.034
- Primary Citation of Related Structures:
2BSF, 2BT7, 2BT8 - PubMed Abstract:
Avian reovirus fibre, a homo-trimer of the sigmaC protein, is responsible for primary host cell attachment. The protein expressed in bacteria forms elongated fibres comprised of a carboxy-terminal globular head domain and a slender shaft, and partial proteolysis yielded a carboxy-terminal protease-stable domain that was amenable to crystallisation. Here, we show that this fragment retains receptor-binding capability and report its structure, solved using two-wavelength anomalous diffraction and refined using data collected from three different crystal forms at 2.1 angstroms, 2.35 angstroms and 3.0 angstroms resolution. The carboxy-terminal globular domain has a beta-barrel fold with the same overall topology as the mammalian reovirus fibre (sigma1). However, the monomers of the sigmaC trimer show a more splayed-out arrangement than in the sigma1 structure. Also resolved are two triple beta-spiral repeats of the shaft or stalk domain. The presence in the sequence of heptad repeats amino-terminal to these triple beta-spiral repeats suggests that the unresolved portion of the shaft domain contains a triple alpha-helical coiled-coil structure. Implications for the function and stability of the sigmaC protein are discussed.
- Departamento de Bioquímica y Biología Molecular, Facultad de Farmacia, Universidad de Santiago de Compostela, Campus Sur, E-15782 Santiago de Compostela, Spain.
Organizational Affiliation: